6EU2
 
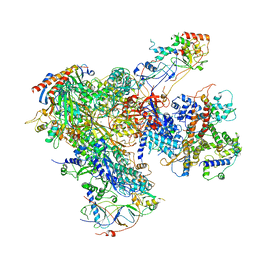 | | Apo RNA Polymerase III - open conformation (oPOL3) | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Abascal-Palacios, G, Ramsay, E.P, Beuron, F, Morris, E, Vannini, A. | | Deposit date: | 2017-10-27 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of RNA polymerase III transcription initiation.
Nature, 553, 2018
|
|
7OBB
 
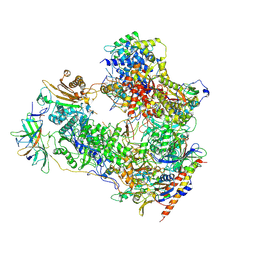 | | Cryo-EM structure of human RNA Polymerase I Open Complex | | Descriptor: | DNA non-template strand, DNA template strand, DNA-directed RNA polymerase I subunit RPA1, ... | | Authors: | Misiaszek, A.D, Girbig, M, Mueller, C.W. | | Deposit date: | 2021-04-21 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of human RNA polymerase I.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7OB9
 
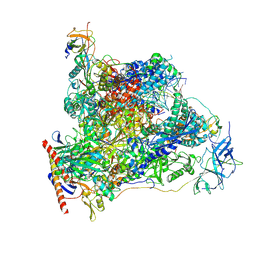 | | Cryo-EM structure of human RNA Polymerase I in elongation state | | Descriptor: | DNA non-template strand, DNA template strand, DNA-directed RNA polymerase I subunit RPA1, ... | | Authors: | Misiaszek, A.D, Girbig, M, Mueller, C.W. | | Deposit date: | 2021-04-21 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of human RNA polymerase I.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7OBA
 
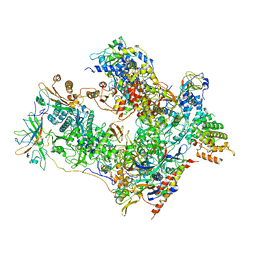 | | Cryo-EM structure of human RNA Polymerase I in complex with RRN3 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA1, DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA2, ... | | Authors: | Misiaszek, A.D, Girbig, M, Mueller, C.W. | | Deposit date: | 2021-04-21 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human RNA polymerase I.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6HKO
 
 | | Yeast RNA polymerase I elongation complex bound to nucleotide analog GMPCPP | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Weis, F, Muller, C.W. | | Deposit date: | 2018-09-07 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2.
Elife, 8, 2019
|
|
6HLR
 
 | | Yeast RNA polymerase I elongation complex bound to nucleotide analog GMPCPP (core focused) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Weis, F, Muller, C.W. | | Deposit date: | 2018-09-11 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2.
Elife, 8, 2019
|
|
6HLS
 
 | | Yeast apo RNA polymerase I* | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Weis, F, Muller, C.W. | | Deposit date: | 2018-09-11 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2.
Elife, 8, 2019
|
|
6HLQ
 
 | | Yeast RNA polymerase I* elongation complex bound to nucleotide analog GMPCPP | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Weis, F, Muller, C.W. | | Deposit date: | 2018-09-11 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2.
Elife, 8, 2019
|
|
7DN3
 
 | | Structure of Human RNA Polymerase III elongation complex | | Descriptor: | DNA (5'-D(P*TP*CP*GP*TP*CP*TP*GP*AP*TP*CP*TP*CP*GP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*CP*CP*GP*AP*GP*AP*TP*CP*AP*GP*AP*CP*GP*AP*GP*AP*TP*CP*GP*GP*G)-3'), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Li, L, Yu, Z, Zhao, D, Ren, Y, Hou, H, Xu, Y. | | Deposit date: | 2020-12-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of human RNA polymerase III elongation complex.
Cell Res., 31, 2021
|
|
7OQ4
 
 | | Cryo-EM structure of the ATV RNAP Inhibitory Protein (RIP) bound to the DNA-binding channel of the host's RNA polymerase | | Descriptor: | Conserved protein, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Pilotto, S, Fouqueau, T, Lukoyanova, N, Sheppard, C, Lucas-Staat, S, Diaz-Santin, L.M, Matelska, D, Prangishvili, D, Cheung, A.C.M, Werner, F. | | Deposit date: | 2021-06-02 | | Release date: | 2021-08-25 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural basis of RNA polymerase inhibition by viral and host factors.
Nat Commun, 12, 2021
|
|
7D58
 
 | | cryo-EM structure of human RNA polymerase III in elongating state | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Wang, Q, Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into transcriptional regulation of human RNA polymerase III.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D59
 
 | | cryo-EM structure of human RNA polymerase III in apo state | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Wang, Q, Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into transcriptional regulation of human RNA polymerase III.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5W66
 
 | | RNA polymerase I Initial Transcribing Complex State 3 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2017-06-16 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
5W5Y
 
 | | RNA polymerase I Initial Transcribing Complex | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2017-06-16 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
5W65
 
 | | RNA polymerase I Initial Transcribing Complex State 2 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2017-06-16 | | Release date: | 2017-08-02 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
5W64
 
 | | RNA Polymerase I Initial Transcribing Complex State 1 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2017-06-16 | | Release date: | 2017-07-26 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
7AOE
 
 | | Schizosaccharomyces pombe RNA polymerase I (elongation complex) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit rpa1, DNA-directed RNA polymerase I subunit rpa14, ... | | Authors: | Heiss, F, Daiss, J, Becker, P, Engel, C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Conserved strategies of RNA polymerase I hibernation and activation.
Nat Commun, 12, 2021
|
|
7AOC
 
 | | Schizosaccharomyces pombe RNA polymerase I (monomer) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit rpa1, DNA-directed RNA polymerase I subunit rpa14, ... | | Authors: | Heiss, F, Daiss, J, Becker, P, Engel, C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Conserved strategies of RNA polymerase I hibernation and activation.
Nat Commun, 12, 2021
|
|
7AOD
 
 | | Schizosaccharomyces pombe RNA polymerase I (dimer) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit rpa1, DNA-directed RNA polymerase I subunit rpa14, ... | | Authors: | Heiss, F, Daiss, J, Becker, P, Engel, C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-02-24 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Conserved strategies of RNA polymerase I hibernation and activation.
Nat Commun, 12, 2021
|
|
6DRD
 
 | | RNA Pol II(G) | | Descriptor: | DNA-directed RNA polymerase II subunit GRINL1A, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Yu, X, Jishage, M, Shi, Y, Ganesan, S, Sali, A, Chait, B.T, Asturias, F, Roeder, R.G. | | Deposit date: | 2018-06-11 | | Release date: | 2018-10-17 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of Pol II(G) and molecular mechanism of transcription regulation by Gdown1.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7OQY
 
 | | Cryo-EM structure of the cellular negative regulator TFS4 bound to the archaeal RNA polymerase | | Descriptor: | Conserved protein, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Pilotto, S, Fouqueau, T, Lukoyanova, N, Sheppard, C, Lucas-Staat, S, Diaz-Santin, L.M, Matelska, D, Prangishvili, D, Cheung, A.C.M, Werner, F. | | Deposit date: | 2021-06-04 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural basis of RNA polymerase inhibition by viral and host factors.
Nat Commun, 12, 2021
|
|
2NVY
 
 | | RNA Polymerase II form II in 150 mM Mn+2 | | Descriptor: | DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, DNA-directed RNA polymerase II 45 kDa polypeptide, ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-13 | | Release date: | 2006-12-26 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
6CNC
 
 | | Yeast RNA polymerase III open complex | | Descriptor: | DNA (71-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
6CNF
 
 | | Yeast RNA polymerase III elongation complex | | Descriptor: | DNA (79-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
7OK0
 
 | | Cryo-EM structure of the Sulfolobus acidocaldarius RNA polymerase at 2.88 A | | Descriptor: | Conserved protein, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Pilotto, S, Fouqueau, T, Lukoyanova, N, Sheppard, C, Lucas-Staat, S, Diaz-Santin, L.M, Matelska, D, Prangishvili, D, Cheung, A.C.M, Werner, F. | | Deposit date: | 2021-05-17 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of RNA polymerase inhibition by viral and host factors.
Nat Commun, 12, 2021
|
|
