5CT8
 
 | | G158E/K44E/R57E/Y49E Bacillus subtilis lipase A with 0% [BMIM][Cl] | | Descriptor: | Quadruple mutant lipase A, SULFATE ION | | Authors: | Nordwald, E.M, Plaks, J.G, Snell, J.R, Sousa, M.C, Kaar, J.L. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystallographic Investigation of Imidazolium Ionic Liquid Effects on Enzyme Structure.
Chembiochem, 16, 2015
|
|
5CTH
 
 | | The 3.7 A resolution structure of a eukaryotic SWEET transporter | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Bidirectional sugar transporter SWEET2b, ... | | Authors: | Feng, L, Tao, Y, Perry, K. | | Deposit date: | 2015-07-24 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.69 Å) | | Cite: | Structure of a eukaryotic SWEET transporter in a homotrimeric complex.
Nature, 527, 2015
|
|
5IKO
 
 | | Crystal structure of human brain glycogen phosphorylase | | Descriptor: | Glycogen phosphorylase, brain form, HEXAETHYLENE GLYCOL, ... | | Authors: | Mathieu, C, Li de la Sierra-Gallay, I, Xu, X, Haouz, A, Rodrigues-Lima, F. | | Deposit date: | 2016-03-03 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into Brain Glycogen Metabolism: THE STRUCTURE OF HUMAN BRAIN GLYCOGEN PHOSPHORYLASE.
J.Biol.Chem., 291, 2016
|
|
8F1I
 
 | | SigN RNA polymerase early-melted intermediate bound to mismatch fragment dhsU36mm1 (-12T) | | Descriptor: | DNA (36-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Mueller, A.U, Chen, J, Darst, S.A. | | Deposit date: | 2022-11-05 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A general mechanism for transcription bubble nucleation in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6U38
 
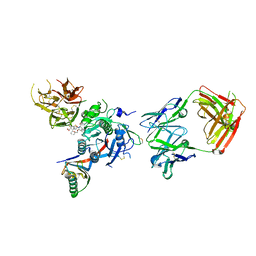 | | PCSK9 in complex with a Fab and compound 8 | | Descriptor: | 2-fluoro-4-{[(1R)-1-methyl-6-{[(2S)-oxan-2-yl]methoxy}-1-{2-oxo-2-[(1,3-thiazol-2-yl)amino]ethyl}-1,2,3,4-tetrahydroisoquinolin-7-yl]oxy}benzoic acid, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Lu, J, Soisson, S. | | Deposit date: | 2019-08-21 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | From Screening to Targeted Degradation: Strategies for the Discovery and Optimization of Small Molecule Ligands for PCSK9.
Cell Chem Biol, 27, 2020
|
|
4HS3
 
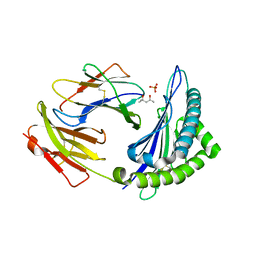 | | Crystal structure of H-2Kb with a disulfide stabilized F pocket in complex with the LCMV derived peptide GP34 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-2-microglobulin, Envelope glycoprotein, ... | | Authors: | Uchtenhagen, H, Boulanger, B, Hein, Z, Abualrous, E.T, Zacharias, M, Werner, J, Elliott, T, Springer, S, Achour, A. | | Deposit date: | 2012-10-29 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Peptide-independent stabilization of MHC class I molecules breaches cellular quality control.
J.Cell.Sci., 127, 2014
|
|
3JC7
 
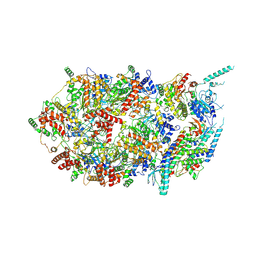 | | Structure of the eukaryotic replicative CMG helicase and pumpjack motion | | Descriptor: | Cell division control protein 45, DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, ... | | Authors: | Li, H, Bai, L, Yuan, Z, Sun, J, Georgescu, R.E, Liu, J, O'Donnell, M.E. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5NP2
 
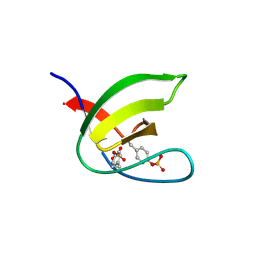 | | Abl1 SH3 pTyr89/134 | | Descriptor: | Tyrosine-protein kinase ABL1 | | Authors: | Mero, B, Radnai, L, Gogl, G, Leveles, I, Buday, L. | | Deposit date: | 2017-04-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the tyrosine phosphorylation-mediated inhibition of SH3 domain-ligand interactions.
J.Biol.Chem., 294, 2019
|
|
3AQ1
 
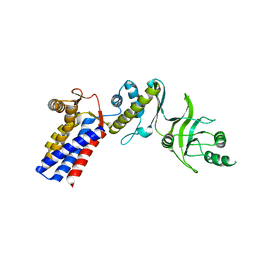 | | Open state monomer of a group II chaperonin from methanococcoides burtonii | | Descriptor: | Thermosome subunit | | Authors: | Harrop, S.J, Pilak, O, Siddiqui, K.S, De Francisci, D, Burg, D, Williams, T.J, Cavicchioli, R, Curmi, P.M. | | Deposit date: | 2010-10-24 | | Release date: | 2011-09-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.746 Å) | | Cite: | Chaperonins from an Antarctic archaeon are predominantly monomeric: crystal structure of an open state monomer.
ENVIRON.MICROBIOL., 13, 2011
|
|
8F1J
 
 | | SigN RNA polymerase early-melted intermediate bound to mismatch DNA fragment dhsU36mm2 (-12A) | | Descriptor: | DNA (36-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Mueller, A.U, Chen, J, Darst, S.A. | | Deposit date: | 2022-11-05 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A general mechanism for transcription bubble nucleation in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8F1K
 
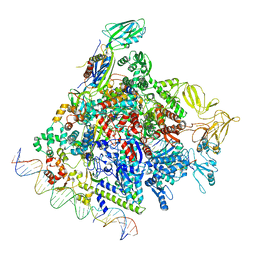 | | SigN RNA polymerase early-melted intermediate bound to full duplex DNA fragment dhsU36 (-12T) | | Descriptor: | DNA (36-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Mueller, A.U, Chen, J, Darst, S.A. | | Deposit date: | 2022-11-05 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A general mechanism for transcription bubble nucleation in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3JAH
 
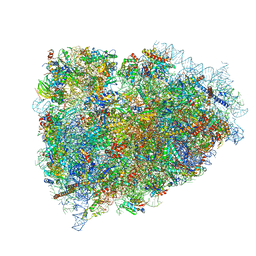 | | Structure of a mammalian ribosomal termination complex with ABCE1, eRF1(AAQ), and the UAG stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Brown, A, Shao, S, Murray, J, Hegde, R.S, Ramakrishnan, V. | | Deposit date: | 2015-06-10 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis for stop codon recognition in eukaryotes.
Nature, 524, 2015
|
|
7N8E
 
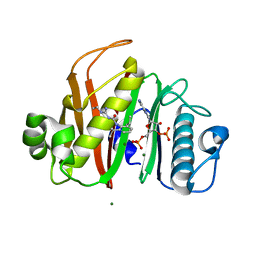 | |
5NPA
 
 | | Solution structure of Drosophila melanogaster Loquacious dsRBD2 | | Descriptor: | Loquacious | | Authors: | Tants, J.-N, Fesser, S, Kern, T, Stehle, R, Geerlof, A, Wunderlich, C, Boettcher, R, Kunzelmann, S, Lange, O, Kreutz, C, Foerstemann, K, Sattler, M. | | Deposit date: | 2017-04-16 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for asymmetry sensing of siRNAs by the Drosophila Loqs-PD/Dcr-2 complex in RNA interference.
Nucleic Acids Res., 45, 2017
|
|
7ZRR
 
 | | Crystal structure of human Urokinase-type plasminogen activator in complex with bicycle peptide inhibitor UK965 | | Descriptor: | 1,2-ETHANEDIOL, 1,3,5-tris(bromomethyl)benzene, AMINO GROUP, ... | | Authors: | Caregnato, A, Angela, P, Mazzoccato, Y, Frasson, N, Angelini, A, Cendron, L. | | Deposit date: | 2022-05-05 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of human Urokinase-type plasminogen activator in complex with bicycle peptide inhibitor UK965
To Be Published
|
|
5NPG
 
 | | Solution structure of Drosophila melanogaster Loquacious dsRBD1 | | Descriptor: | Loquacious, isoform F | | Authors: | Tants, J.-N, Fesser, S, Kern, T, Stehle, R, Geerlof, A, Wunderlich, C, Hartlmuller, C, Boettcher, R, Kunzelmann, S, Lange, O, Kreutz, C, Foerstemann, K, Sattler, M. | | Deposit date: | 2017-04-16 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for asymmetry sensing of siRNAs by the Drosophila Loqs-PD/Dcr-2 complex in RNA interference.
Nucleic Acids Res., 45, 2017
|
|
7V3X
 
 | |
5ILI
 
 | | Tobacco 5-epi-aristolochene synthase with CAPSO buffer molecule and Mg2+ ions | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 5-epi-aristolochene synthase, ... | | Authors: | Koo, H.J, Louie, G.V, Xu, Y, Bowman, M, Noel, J.P. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small-molecule buffer components can directly affect terpene-synthase activity by interacting with the substrate-binding site of the enzyme
To Be Published
|
|
4HZT
 
 | | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates | | Descriptor: | 3-{(1S)-1-[(6-chloro-3,3-dimethyl-3,4-dihydroisoquinolin-1-yl)amino]-2-phenylethyl}-1,2,4-oxadiazol-5(2H)-one, Beta-secretase 1, ZINC ION | | Authors: | Yao, N, Brecht, E. | | Deposit date: | 2012-11-15 | | Release date: | 2013-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of novel dihydroisoquinoline BACE-1 inhibitors that do not engage the catalytic aspartates.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4I03
 
 | | Human MMP12 in complex with a PEG-linked bifunctional L-glutamate motif inhibitor | | Descriptor: | (4R,22R)-5,21-dioxo-4,22-bis({3-[4-(4-phenylthiophen-2-yl)phenyl]propanoyl}amino)-10,13,16-trioxa-6,20-diazapentacosane-1,25-dioic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Stura, E.A, Vera, L, Devel, L, Cassar-Lajeunesse, E, Dive, V. | | Deposit date: | 2012-11-16 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallization of bi-functional ligand protein complexes.
J.Struct.Biol., 182, 2013
|
|
5UCO
 
 | |
5IEB
 
 | |
4I06
 
 | | Crystal structure of human Arginase-2 complexed with inhibitor 14 | | Descriptor: | Arginase-2, mitochondrial, BENZAMIDINE, ... | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Whitehouse, D.L, Golebiowski, A, Ji, M, Zhang, M, Beckett, P, Sheeler, R, Andreoli, M, Conway, B, Mahboubi, K, Schroeter, H, Van Zandt, M.C, Podjarny, A. | | Deposit date: | 2012-11-16 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of (R)-2-Amino-6-borono-2-(2-(piperidin-1-yl)ethyl)hexanoic Acid and Congeners As Highly Potent Inhibitors of Human Arginases I and II for Treatment of Myocardial Reperfusion Injury.
J.Med.Chem., 56, 2013
|
|
4I0Y
 
 | |
6U7M
 
 | |
