5GUS
 
 | | Crystal structure of ASCH domain from Zymomonas mobilis | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CHLORIDE ION, Helix-turn-helix domain-containing protein, ... | | Authors: | Ha, S.C, Park, S.Y, Kim, J.S. | | Deposit date: | 2016-08-31 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Crystal structure of an ASCH protein from Zymomonas mobilis and its ribonuclease activity specific for single-stranded RNA.
Sci Rep, 7, 2017
|
|
8XJ2
 
 | | MAT2A in complex with SAM and allosteric inhibitor SCR-7952 | | Descriptor: | 7-chloranyl-5-(2-cyclopropylpyridin-3-yl)-1-methyl-imidazo[4,5-c]quinolin-4-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Zhou, F. | | Deposit date: | 2023-12-20 | | Release date: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | MAT2A in complex with SAM and allosteric inhibitor SCR-7952
To Be Published
|
|
5UTU
 
 | | 2.65 Angstrom Resolution Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with SAH and NAD | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Stam, J, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 2.65 Angstrom Resolution Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with SAH and NAD
To Be Published
|
|
6IPB
 
 | |
4H2M
 
 | | Structure of E. coli undecaprenyl diphosphate synthase in complex with BPH-1408 | | Descriptor: | 2,2'-{benzene-1,3-diylbis[ethyne-2,1-diyl(5-bromobenzene-3,1-diyl)]}diethanamine, Undecaprenyl pyrophosphate synthase | | Authors: | Zhu, W, Oldfield, E. | | Deposit date: | 2012-09-12 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Antibacterial drug leads targeting isoprenoid biosynthesis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8PSW
 
 | | ERK2 covalently bound to RU67 cyclohexenone based inhibitor | | Descriptor: | AMP PHOSPHORAMIDATE, Mitogen-activated protein kinase 1, ~{O}3-~{tert}-butyl ~{O}1-methyl (1~{R},3~{R})-4-oxidanylidene-1-(phenylmethyl)cyclohexane-1,3-dicarboxylate | | Authors: | Sok, P, Poti, A, Remenyi, A, Gogl, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a precision-guided warhead scaffold.
Nat Commun, 15, 2024
|
|
8XJF
 
 | | Crystal structure of Arabidopsis N-amino acetyltransferase 2 bound to HEPES, Acetyl CoA, GABA, and glycerol | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYL COENZYME *A, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Arold, S.T, Hameed, U.F.S. | | Deposit date: | 2023-12-21 | | Release date: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Arabidopsis N-amino acetyltransferase 2 bound to HEPES, Acetyl CoA, GABA, and glycerol
To Be Published
|
|
6V1K
 
 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to BI7273 | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-[(dimethylamino)methyl]-3,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, Bromodomain-containing protein 4 | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
8PT1
 
 | | ERK2 covelently bound to RU76 cyclohexenone based inhibitor | | Descriptor: | Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ~{O}1-methyl ~{O}3-(phenylmethyl) (1~{S},3~{R})-1-methyl-4-oxidanylidene-cyclohexane-1,3-dicarboxylate | | Authors: | Sok, P, Poti, A, Remenyi, A, Gogl, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a precision-guided warhead scaffold.
Nat Commun, 15, 2024
|
|
9MMQ
 
 | | Cryo-EM structure of CRAF/MEK1 complex (kinase domain) | | Descriptor: | 5-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)imidazo[1,5-a]pyridine-6-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Jang, D.M, Jeon, H, Eck, M.J. | | Deposit date: | 2024-12-20 | | Release date: | 2025-09-10 | | Last modified: | 2025-10-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of CRAF/MEK1/14-3-3 complexes in autoinhibited and open-monomer states reveal features of RAF regulation.
Nat Commun, 16, 2025
|
|
3BH8
 
 | | Crystal Structure of RQA_M Phosphopeptide Bound to HUMAN Class I MHC HLA-A2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Mohammed, F, Cobbold, M, Zarling, A.L, Salim, M, Barrett-Wilt, G.A, Shabanowitz, J, Hunt, D.F, Engelhard, V.H, Willcox, B.E. | | Deposit date: | 2007-11-28 | | Release date: | 2008-10-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self
Nat.Immunol., 9, 2008
|
|
9E42
 
 | | RTA-RUNT-192 complex | | Descriptor: | 1,2-ETHANEDIOL, 5-(2,6-diethylphenyl)thiophene-2-carboxylic acid, Ricin A chain | | Authors: | Rudolph, M.J, Tumer, N. | | Deposit date: | 2024-10-24 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of small molecules at the P-stalk site of ricin A subunit trigger conformational changes that extend into the active site.
J.Biol.Chem., 301, 2025
|
|
6IVD
 
 | | TGEV nsp1 mutant - 91-95sg | | Descriptor: | nsp1 mutant protein | | Authors: | Shen, Z, Peng, G.Q. | | Deposit date: | 2018-12-03 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | A conserved region of nonstructural protein 1 from alphacoronaviruses inhibits host gene expression and is critical for viral virulence.
J.Biol.Chem., 294, 2019
|
|
6IVM
 
 | | Crystal structure of a membrane protein P143A | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
6IVX
 
 | | Discovery of the Second Generation ROR gamma Inhibitors Composed of an Azole Scaffold. | | Descriptor: | (4S)-4-[4'-cyclopropyl-5-(2,2-dimethylpropyl)[3,5'-bi-1,2-oxazol]-3'-yl]-6-[(2,4-dichlorophenyl)amino]-6-oxohexanoic acid, Nuclear receptor ROR-gamma, Nuclear receptor corepressor 2 | | Authors: | Noguchi, M, Nomura, A, Doi, S, Adachi, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Second Generation ROR gamma Inhibitors Composed of an Azole Scaffold.
J. Med. Chem., 62, 2019
|
|
5V8T
 
 | | Crystal structure of SMT fusion Peptidyl-prolyl cis-trans isomerase from Burkholderia pseudomallei complexed with SF354 | | Descriptor: | 2-{[3,5-bis(2-methoxyethoxy)benzene-1-carbonyl]amino}ethyl (2S)-1-(benzylsulfonyl)piperidine-2-carboxylate, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Lorimer, D.D, Dranow, D.M, Seufert, F, Abendroth, J, Holzgrabe, U. | | Deposit date: | 2017-03-22 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of SMT fusion Peptidyl-prolyl cis-trans isomerase from Burkholderia pseudomallei complexed with SF354
to be published
|
|
8PT3
 
 | | ERK2 covelently bound to RU77 cyclohexenone based inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Sok, P, Poti, A, Remenyi, A. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a precision-guided warhead scaffold.
Nat Commun, 15, 2024
|
|
6I5D
 
 | | Crystal structure of an OXA-48 beta-lactamase synthetic mutant | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Zavala, A, Retailleau, P, Dabos, L, Naas, T, Iorga, B. | | Deposit date: | 2018-11-13 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate specificity of an OXA-48 beta-lactamase synthetic mutant
To be published
|
|
9NFB
 
 | |
4YZ3
 
 | | Crystal Structure of Streptococcus pneumoniae NanC, in complex with Oseltamivir. | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, Neuraminidase, SULFATE ION | | Authors: | Lukacik, P, Owen, C.D, Potter, J.A, Taylor, G.L, Walsh, M.A. | | Deposit date: | 2015-03-24 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of Streptococcus pneumoniae NanC.
To Be Published
|
|
5V9I
 
 | | Crystal structure of catalytic domain of G9a with MS0105 | | Descriptor: | GLYCEROL, Histone-lysine N-methyltransferase EHMT2, N~2~-cyclohexyl-N~4~-(1-ethylpiperidin-4-yl)-6,7-dimethoxy-N~2~-methylquinazoline-2,4-diamine, ... | | Authors: | Dong, A, Zeng, H, Liu, J, Xiong, Y, Babault, N, Jin, J, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Wu, H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-23 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of catalytic domain of G9a with MS0105
to be published
|
|
1WS1
 
 | |
8PST
 
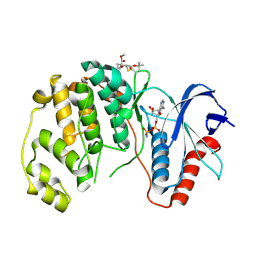 | | ERK2 covelently bound to RU60 cyclohexenone based inhibitor | | Descriptor: | AMP PHOSPHORAMIDATE, Mitogen-activated protein kinase 1, ~{O}3-~{tert}-butyl ~{O}1-methyl (1~{R},3~{S})-1-methyl-4-oxidanylidene-cyclohexane-1,3-dicarboxylate | | Authors: | Sok, P, Poti, A, Remenyi, A, Gogl, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a precision-guided warhead scaffold.
Nat Commun, 15, 2024
|
|
7LCQ
 
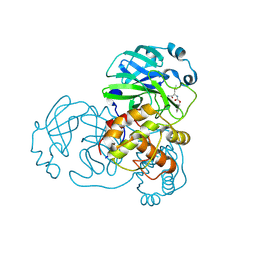 | | N-terminal finger stabilizes feline drug GC376 in coronavirus 3CL protease | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Khan, M.B, Arutyunova, E, Young, H.S, Lemieux, M.J. | | Deposit date: | 2021-01-11 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | N-Terminal Finger Stabilizes the S1 Pocket for the Reversible Feline Drug GC376 in the SARS-CoV-2 M pro Dimer.
J.Mol.Biol., 433, 2021
|
|
5VBS
 
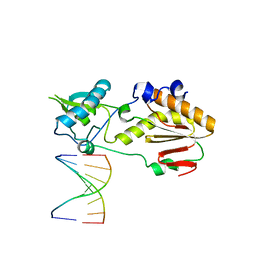 | | Structural basis for a six letter alphabet including GATCKX | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*TP*(DX)P*(DX)P*T)-3'), DNA (5'-D(P*AP*(93D)P*(93D)P*AP*TP*AP*AP*G)-3'), reverse transcriptase catalytic fragment | | Authors: | Singh, I, Georgiadis, M.M. | | Deposit date: | 2017-03-30 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Structure and Biophysics for a Six Letter DNA Alphabet that Includes Imidazo[1,2-a]-1,3,5-triazine-2(8H)-4(3H)-dione (X) and 2,4-Diaminopyrimidine (K).
ACS Synth Biol, 6, 2017
|
|
