5TT7
 
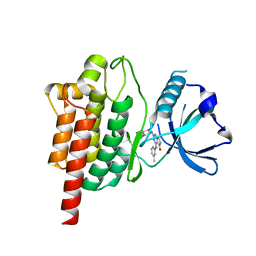 | | Discovery of TAK-659, an Orally Available Investigational Inhibitor of Spleen Tyrosine Kinase (SYK) | | Descriptor: | 2-{[(1R,2S)-2-aminocyclohexyl]amino}-4-[(3-methylphenyl)amino]-6,7-dihydro-5H-pyrrolo[3,4-d]pyrimidin-5-one, Tyrosine-protein kinase SYK | | Authors: | Yano, J, Jennings, A, Lam, B, Hoffman, I.D. | | Deposit date: | 2016-11-01 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Discovery of TAK-659 an orally available investigational inhibitor of Spleen Tyrosine Kinase (SYK).
Bioorg. Med. Chem. Lett., 26, 2016
|
|
5TVC
 
 | | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 3 nicotinic acetylcholine receptor in complex with (E,2S)-N-methyl-5-(5-phenoxy-3-pyridyl)pent-4-en-2-amine (TI-5312) | | Descriptor: | (E,2S)-N-methyl-5-(5-phenoxy-3-pyridyl)pent-4-en-2-amine, PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Bobango, J, Wu, J, Talley, I.T, Talley, T.T. | | Deposit date: | 2016-11-08 | | Release date: | 2016-11-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.926 Å) | | Cite: | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 3 nicotinic acetylcholine receptor in complex with (E,2S)-N-methyl-5-(5-phenoxy-3-pyridyl)pent-4-en-2-amine (TI-5312)
To Be Published
|
|
5WHD
 
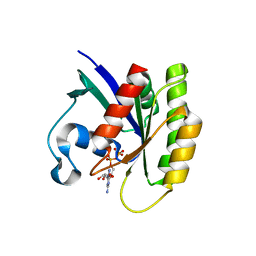 | | Crystal structure of KRas G12V/D38P, bound to GDP | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Shim, S.Y, McGee, J.H, Lee, S.-J, Verdine, G.L. | | Deposit date: | 2017-07-16 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Exceptionally high-affinity Ras binders that remodel its effector domain.
J. Biol. Chem., 293, 2018
|
|
5WCI
 
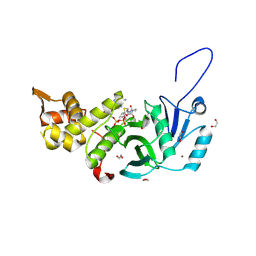 | | Human MYST histone acetyltransferase 1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Histone acetyltransferase KAT8, ... | | Authors: | Dong, A, Zeng, H, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Zheng, Y.G, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-30 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Human MYST histone acetyltransferase 1
to be published
|
|
5WMR
 
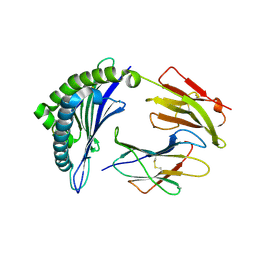 | | Crystal Structure of HLA-B8 in complex with QIK, a CMV peptide | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-8 alpha chain, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-31 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Inability To Detect Cross-Reactive Memory T Cells Challenges the Frequency of Heterologous Immunity among Common Viruses.
J. Immunol., 200, 2018
|
|
7LZS
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-11029 | | Descriptor: | 3-chloro-5-{7-[2-({5-chloro-2-[(3S)-3-methylmorpholin-4-yl]pyridin-4-yl}amino)-2-oxoethyl]-3-methyl-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl}-2-hydroxybenzamide, B-cell lymphoma 6 protein, DIMETHYL SULFOXIDE, ... | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of OICR12694: A Novel, Potent, Selective, and Orally Bioavailable BCL6 BTB Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
5WP6
 
 | | Cryo-EM structure of a human TRPM4 channel in complex with calcium and decavanadate | | Descriptor: | DECAVANADATE, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Winkler, P.A, Huang, Y, Sun, W, Du, J, Lu, W. | | Deposit date: | 2017-08-03 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Electron cryo-microscopy structure of a human TRPM4 channel.
Nature, 552, 2017
|
|
5WJN
 
 | | Crystal Structure of HLA-A*11:01-GTS3 | | Descriptor: | Beta-2-microglobulin, GTS3 peptide, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-23 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8(+) T cell response.
Nat. Immunol., 18, 2017
|
|
5WQU
 
 | | Crystal structure of Sweet Potato Beta-Amylase complexed with Maltotetraose | | Descriptor: | Beta-amylase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Vajravijayan, S, Sergei, P, Nandhagopal, N, Gunasekaran, K. | | Deposit date: | 2016-11-28 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural insights on starch hydrolysis by plant beta-amylase and its evolutionary relationship with bacterial enzymes
Int. J. Biol. Macromol., 113, 2018
|
|
5W7Q
 
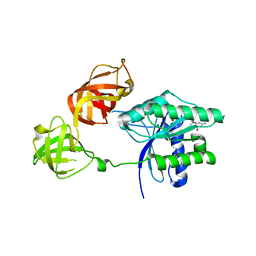 | |
5A0Z
 
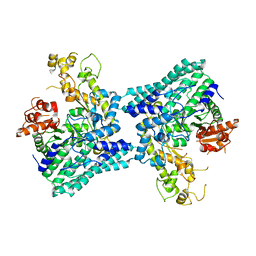 | |
7N1F
 
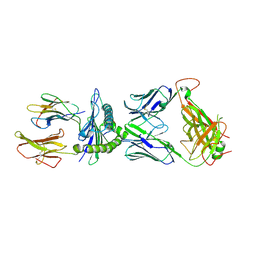 | | SARS-CoV-2 YLQ peptide-specific TCR pYLQ7 binds to YLQ-HLA-A2 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, A-2 alpha chain, ... | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
Nat Commun, 13, 2022
|
|
7N65
 
 | | Complex structure of HIV superinfection Fab QA013.2 and BG505.SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp41, ... | | Authors: | Mangala Prasad, V, Shipley, M.M, Overbaugh, J.M, Lee, K.K. | | Deposit date: | 2021-06-07 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Functional development of a V3/glycan-specific broadly neutralizing antibody isolated from a case of HIV superinfection.
Elife, 10, 2021
|
|
5A28
 
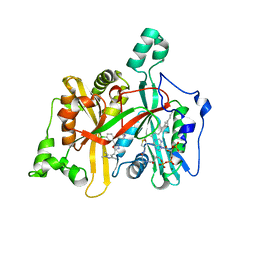 | | Leishmania major N-myristoyltransferase in complex with a chlorophenyl 1,3,4-oxadiazole inhibitor. | | Descriptor: | 4-(4-chloro-2-{5-[(trimethyl-1H-pyrazol-4-yl)methyl]-1,3,4-oxadiazol-2-yl}phenoxy)piperidine, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Rackham, M.D, Yu, Z, Brannigan, J.A, Heal, W.P, Paape, D, Barker, K.V, Wilkinson, A.J, Smith, D.F, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2015-05-15 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Discovery of High Affinity Inhibitors of Leishmania Donovani N-Myristoyltransferase.
Medchemcomm, 6, 2015
|
|
5WBM
 
 | |
5WBR
 
 | | Structure of human Ketohexokinase complexed with hits from fragment screening | | Descriptor: | 6-[4-(2-hydroxyethyl)piperazin-1-yl]-2-[(3S)-3-(hydroxymethyl)piperidin-1-yl]-4-(trifluoromethyl)pyridine-3-carbonitrile, CITRIC ACID, GLYCEROL, ... | | Authors: | Pandit, J. | | Deposit date: | 2017-06-29 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of Fragment-Derived Small Molecules for in Vivo Inhibition of Ketohexokinase (KHK).
J. Med. Chem., 60, 2017
|
|
5AHF
 
 | | Crystal structure of Salmonella enterica HisA D7N with ProFAR | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, GLYCEROL, [(2R,3S,4R,5R)-5-[4-aminocarbonyl-5-[(E)-[[(2R,3R,4S,5R)-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]amino]methylideneamino]imidazol-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Soderholm, A, Guo, X, Newton, M.S, Evans, G.B, Nasvall, J, Patrick, W.M, Selmer, M. | | Deposit date: | 2015-02-05 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Two-Step Ligand Binding in a Beta/Alpha8 Barrel Enzyme -Substrate-Bound Structures Shed New Light on the Catalytic Cycle of Hisa
J.Biol.Chem., 290, 2015
|
|
5WDK
 
 | | A processive dipeptidyl aminopeptidase secreted from an established commensal bacterium P. distasonis | | Descriptor: | 5-[(3aS,4R,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-N-(2-oxopropyl)pentanamide, Aminopeptidase C, POTASSIUM ION | | Authors: | Wolan, D.W, Xu, J.H, Solania, A, Chatterjee, S, Jiang, Z, ODonoghue, A.J. | | Deposit date: | 2017-07-05 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | A Commensal Dipeptidyl Aminopeptidase with Specificity for N-Terminal Glycine Degrades Human-Produced Antimicrobial Peptides in Vitro.
Acs Chem.Biol., 13, 2018
|
|
5WEV
 
 | |
5V75
 
 | |
5VC4
 
 | | Crystal structure of HUMAN WEE1 KINASE domain in complex with Bosutinib-isomer | | Descriptor: | 4-[(3,5-DICHLORO-4-METHOXYPHENYL)AMINO]-6-METHOXY-7-[3-(4-METHYLPIPERAZIN-1-YL)PROPOXY]QUINOLINE-3-CARBONITRILE, PHOSPHATE ION, Wee1-like protein kinase | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-03-30 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Wee Kinases Functionality and Inactivation by Diverse Small Molecule Inhibitors.
J. Med. Chem., 60, 2017
|
|
5A4M
 
 | | Mechanism of Hydrogen activation by NiFe-hydrogenases | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Evans, R.M, Brooke, E.J, Wehlin, S.A.M, Nomerotskaia, E, Sergent, F, Carr, S.B, Philips, S.E.V, Armstrong, F.A. | | Deposit date: | 2015-06-10 | | Release date: | 2015-11-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Hydrogen Activation by [Nife] Hydrogenases.
Nat.Chem.Biol., 12, 2016
|
|
7MYL
 
 | |
5UK6
 
 | | Structure of Anabaena Sensory Rhodopsin Determined by Solid State NMR Spectroscopy and DEER | | Descriptor: | Bacteriorhodopsin | | Authors: | Milikisiyants, S, Wang, S, Munro, R.A, Donohue, M, Ward, M.E, Brown, L.S, Smirnova, T.I, Ladizhansky, V, Smirnov, A.I. | | Deposit date: | 2017-01-20 | | Release date: | 2017-05-31 | | Last modified: | 2020-01-08 | | Method: | SOLID-STATE NMR | | Cite: | Oligomeric Structure of Anabaena Sensory Rhodopsin in a Lipid Bilayer Environment by Combining Solid-State NMR and Long-range DEER Constraints.
J. Mol. Biol., 429, 2017
|
|
5AHO
 
 | | Crystal structure of human 5' exonuclease Apollo | | Descriptor: | 1,2-ETHANEDIOL, 5' EXONUCLEASE APOLLO, L(+)-TARTARIC ACID, ... | | Authors: | Allerston, C.K, Vollmar, M, Krojer, T, Pike, A.C.W, Newman, J.A, Carpenter, E, Quigley, A, Mahajan, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Gileadi, O. | | Deposit date: | 2015-02-06 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The Structures of the Snm1A and Snm1B/Apollo Nuclease Domains Reveal a Potential Basis for Their Distinct DNA Processing Activities.
Nucleic Acids Res., 43, 2015
|
|
