2JEF
 
 | | The Molecular Basis of Selectivity of Nucleotide Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus solfataricus DNA Polymerase IV: Steady-state and Pre-steady-state and X-Ray Crystallography of Correct and Incorrect Pairing | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*CP*DOC)-3', 5'-D(*TP*CP*AP*CP*BZGP*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP CP*CP*C)-3', ... | | Authors: | Eoff, R.L, Angel, K.C, Kosekov, I.D, Egli, M, Guengerich, F.P. | | Deposit date: | 2007-01-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus Solfataricus DNA Polymerase Dpo4: Steady-State and Pre-Steady-State Kinetics and X-Ray Crystallography of Correct and Incorrect Pairing.
J.Biol.Chem., 282, 2007
|
|
2JEG
 
 | | The Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus solfataricus DNA Polymerase IV: Steady-state and Pre-steady-state Kinetics and X- Ray Crystallography of Correct and Incorrect Pairing | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*DOC)-3', 5'-D(*TP*CP*AP*C BZGP*GP*AP*AP*TP*CP *CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Eoff, R.L, Angel, K.C, Kosekov, I.D, Egli, M, Guengerich, F.P. | | Deposit date: | 2007-01-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus Solfataricus DNA Polymerase Dpo4: Steady-State and Pre-Steady-State Kinetics and X-Ray Crystallography of Correct and Incorrect Pairing.
J.Biol.Chem., 282, 2007
|
|
2JEI
 
 | | The Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus solfataricus DNA Polymerase IV: Steady-state and Pre-steady-state Kinetics and X- Ray Crystallography of Correct and Incorrect Pairing | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*T)-3', 5'-D(*TP*CP*AP*C BZGP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Eoff, R.L, Angel, K.C, Kosekov, I.D, Egli, M, Guengerich, F.P. | | Deposit date: | 2007-01-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus Solfataricus DNA Polymerase Dpo4: Steady-State and Pre-Steady-State Kinetics and X-Ray Crystallography of Correct and Incorrect Pairing.
J.Biol.Chem., 282, 2007
|
|
2JEJ
 
 | | The Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus solfataricus DNA Polymerase IV: Steady-state and Pre-steady-state Kinetics and X- Ray Crystallography of Correct and Incorrect Pairing | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*CP*G)-3', 5'-D(*TP*CP*AP*C BZGP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Eoff, R.L, Angel, K.C, Kosekov, I.D, Egli, M, Guengerich, F.P. | | Deposit date: | 2007-01-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Molecular Basis of Selectivity of Nucleoside Triphosphate Incorporation Opposite O6-Benzylguanine by Sulfolobus Solfataricus DNA Polymerase Dpo4: Steady-State and Pre-Steady-State Kinetics and X-Ray Crystallography of Correct and Incorrect Pairing.
J.Biol.Chem., 282, 2007
|
|
2JEK
 
 | | Crystal structure of the conserved hypothetical protein Rv1873 from Mycobacterium tuberculosis at 1.38 A | | Descriptor: | GLYCEROL, RV1873, SULFATE ION | | Authors: | Garen, C.R, Cherney, M.M, Bergmann, E.M, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-01-17 | | Release date: | 2007-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The Molecular Structure of Rv1873, a Conserved Hypothetical Protein from Mycobacterium Tuberculosis, at 1.38A Resolution
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2JEL
 
 | | JEL42 FAB/HPR COMPLEX | | Descriptor: | HISTIDINE-CONTAINING PROTEIN, JEL42 FAB FRAGMENT, SULFATE ION | | Authors: | Prasad, L, Waygood, E.B, Lee, J.S, Delbaere, L.T.J. | | Deposit date: | 1998-02-24 | | Release date: | 1998-05-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 2.5 A resolution structure of the jel42 Fab fragment/HPr complex
J.Mol.Biol., 280, 1998
|
|
2JEM
 
 | | Native family 12 xyloglucanase from Bacillus licheniformis | | Descriptor: | ENDO-BETA-1,4-GLUCANASE | | Authors: | Gloster, T.M, Ibatullin, F.M, Macauley, K, Eklof, J.M, Roberts, S, Turkenburg, J.P, Bjornvad, M.E, Jorgensen, P.L, Danielsen, S, Johansen, K.S, Borchert, T.V, Wilson, K.S, Brumer, H, Davies, G.J. | | Deposit date: | 2007-01-18 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Characterization and Three-Dimensional Structures of Two Distinct Bacterial Xyloglucanases from Families Gh5 and Gh12.
J.Biol.Chem., 282, 2007
|
|
2JEN
 
 | | Family 12 xyloglucanase from Bacillus licheniformis in complex with ligand | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ENDO-BETA-1,4-GLUCANASE, GLYCEROL, ... | | Authors: | Gloster, T.M, Ibatullin, F.M, Macauley, K, Eklof, J.M, Roberts, S, Turkenburg, J.P, Bjornvad, M.E, Jorgensen, P.L, Danielsen, S, Johansen, K.S, Borchert, T.V, Wilson, K.S, Brumer, H, Davies, G.J. | | Deposit date: | 2007-01-18 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization and Three-Dimensional Structures of Two Distinct Bacterial Xyloglucanases from Families Gh5 and Gh12.
J.Biol.Chem., 282, 2007
|
|
2JEO
 
 | | Crystal structure of human uridine-cytidine kinase 1 | | Descriptor: | URIDINE-CYTIDINE KINASE 1 | | Authors: | Kosinska, U, Stenmark, P, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Hogbom, M, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E.P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Uppsten, M, Thorsell, A.G, van den Berg, S, Weigelt, J, Welin, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Human Uridine-Cytidine Kinase 1
To be Published
|
|
2JEP
 
 | | Native family 5 xyloglucanase from Paenibacillus pabuli | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, XYLOGLUCANASE | | Authors: | Gloster, T.M, Ibatullin, F.M, Macauley, K, Eklof, J.M, Roberts, S, Turkenburg, J.P, Bjornvad, M.E, Jorgensen, P.L, Danielsen, S, Johansen, K, Borchert, T.V, Wilson, K.S, Brumer, H, Davies, G.J. | | Deposit date: | 2007-01-18 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization and Three-Dimensional Structures of Two Distinct Bacterial Xyloglucanases from Families Gh5 and Gh12.
J.Biol.Chem., 282, 2007
|
|
2JEQ
 
 | | Family 5 xyloglucanase from Paenibacillus pabuli in complex with ligand | | Descriptor: | XYLOGLUCANASE, beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[beta-D-galactopyranose-(1-2)-alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Gloster, T.M, Ibatullin, F.M, Macauley, K, Eklof, J.M, Roberts, S, Turkenburg, J.P, Bjornvad, M.E, Jorgensen, P.L, Danielsen, S, Johansen, K, Borchert, T.V, Wilson, K.S, Brumer, H, Davies, G.J. | | Deposit date: | 2007-01-18 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Characterization and Three-Dimensional Structures of Two Distinct Bacterial Xyloglucanases from Families Gh5 and Gh12.
J.Biol.Chem., 282, 2007
|
|
2JER
 
 | |
2JES
 
 | | Portal protein (gp6) from bacteriophage SPP1 | | Descriptor: | CALCIUM ION, MERCURY (II) ION, PORTAL PROTEIN, ... | | Authors: | Lebedev, A.A, Krause, M.H, Isidro, A.L, Vagin, A.A, Orlova, E.V, Turner, J, Dodson, E.J, Tavares, P, Antson, A.A. | | Deposit date: | 2007-01-21 | | Release date: | 2007-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Framework for DNA Translocation Via the Viral Portal Protein
Embo J., 26, 2007
|
|
2JET
 
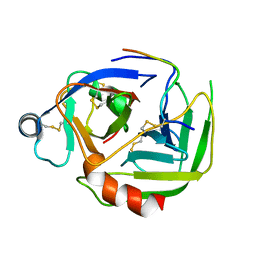 | | Crystal structure of a trypsin-like mutant (S189D , A226G) chymotrypsin. | | Descriptor: | CHYMOTRYPSINOGEN B CHAIN A, CHYMOTRYPSINOGEN B CHAIN B, CHYMOTRYPSINOGEN B CHAIN C | | Authors: | Jelinek, B, Katona, G, Fodor, K, Venekei, I, Graf, L. | | Deposit date: | 2007-01-22 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of a Trypsin-Like Mutant Chymotrypsin: The Role of Position 226 in the Activity and Specificity of S189D Chymotrypsin.
Protein J., 27, 2008
|
|
2JEU
 
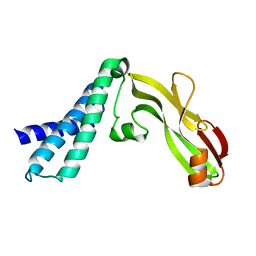 | | Transcription activator structure reveals redox control of a replication initiation reaction | | Descriptor: | REGULATORY PROTEIN E2 | | Authors: | Sanders, C.M, Sizov, D, Seavers, P.R, Ortiz-Lombardia, M, Antson, A.A. | | Deposit date: | 2007-01-23 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Transcription Activator Structure Reveals Redox Control of a Replication Initiation Reaction.
Nucleic Acids Res., 35, 2007
|
|
2JEV
 
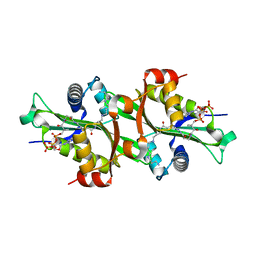 | | Crystal structure of human spermine,spermidine acetyltransferase in complex with a bisubstrate analog (N1-acetylspermine-S-CoA). | | Descriptor: | (3R)-27-AMINO-3-HYDROXY-2,2-DIMETHYL-4,8,14-TRIOXO-12-THIA-5,9,15,19,24-PENTAAZAHEPTACOS-1-YL [(2S,3R,4S,5S)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE, DIAMINE ACETYLTRANSFERASE 1 | | Authors: | Hegde, S.S, Chandler, J, Vetting, M.W, Yu, M, Blanchard, J.S. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic and Structural Analysis of Human Spermidine/Spermine N(1)-Acetyltransferase.
Biochemistry, 46, 2007
|
|
2JEW
 
 | |
2JEX
 
 | | Transcription activator structure reveals redox control of a replication initiation reaction | | Descriptor: | REGULATORY PROTEIN E2 | | Authors: | Sanders, C.M, Sizov, D, Seavers, P.R, Ortiz-Lombardia, M, Antson, A.A. | | Deposit date: | 2007-01-24 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Transcription Activator Structure Reveals Redox Control of a Replication Initiation Reaction.
Nucleic Acids Res., 35, 2007
|
|
2JEY
 
 | | Mus musculus acetylcholinesterase in complex with HLo-7 | | Descriptor: | 1-[({2,4-BIS[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]-4-CARBAMOYLPYRIDINIUM, ACETYLCHOLINESTERASE, HEXAETHYLENE GLYCOL | | Authors: | Ekstrom, F, Astot, C, Pang, Y.P. | | Deposit date: | 2007-01-25 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Novel Nerve-Agent Antidote Design Based on Crystallographic and Mass Spectrometric Analyses of Tabun-Conjugated Acetylcholinesterase in Complex with Antidotes.
Clin.Pharmacol.Ther., 82, 2007
|
|
2JEZ
 
 | | Mus musculus acetylcholinesterase in complex with tabun and HLo-7 | | Descriptor: | 1-[({2,4-BIS[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]-4-CARBAMOYLPYRIDINIUM, ACETYLCHOLINESTERASE, HEXAETHYLENE GLYCOL | | Authors: | Ekstrom, F, Astot, C, Pang, Y.P. | | Deposit date: | 2007-01-25 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel Nerve-Agent Antidote Design Based on Crystallographic and Mass Spectrometric Analyses of Tabun-Conjugated Acetylcholinesterase in Complex with Antidotes.
Clin.Pharmacol.Ther., 82, 2007
|
|
2JF0
 
 | | Mus musculus acetylcholinesterase in complex with tabun and Ortho-7 | | Descriptor: | 1,7-HEPTYLENE-BIS-N,N'-SYN-2-PYRIDINIUMALDOXIME, ACETYLCHOLINESTERASE, HEXAETHYLENE GLYCOL | | Authors: | Ekstrom, F, Astot, C, Pang, Y.P. | | Deposit date: | 2007-01-25 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel Nerve-Agent Antidote Design Based on Crystallographic and Mass Spectrometric Analyses of Tabun-Conjugated Acetylcholinesterase in Complex with Antidotes.
Clin.Pharmacol.Ther., 82, 2007
|
|
2JF1
 
 | |
2JF2
 
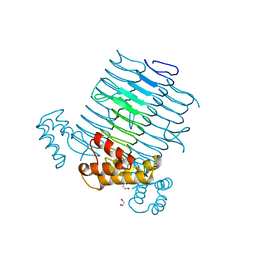 | | Nucleotide substrate binding by UDP-N-acetylglucosamine acyltransferase | | Descriptor: | 1,2-ETHANEDIOL, ACYL-[ACYL-CARRIER-PROTEIN]--UDP-N-ACETYLGLUCOSAMINE O-ACYLTRANSFERASE, DI(HYDROXYETHYL)ETHER | | Authors: | Ulaganathan, V, Buetow, L, Hunter, W.N. | | Deposit date: | 2007-01-25 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nucleotide Substrate Recognition by Udp-N-Acetylglucosamine Acyltransferase (Lpxa) in the First Step of Lipid a Biosynthesis.
J.Mol.Biol., 369, 2007
|
|
2JF3
 
 | |
2JF4
 
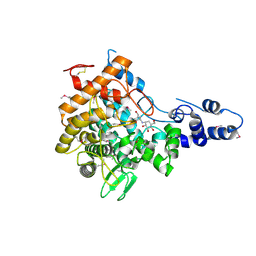 | | Family 37 trehalase from Escherichia coli in complex with validoxylamine | | Descriptor: | (1S,2S,3R,6S)-4-(HYDROXYMETHYL)-6-{[(1S,2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-(HYDROXYMETHYL)CYCLOHEXYL]AMINO}CYCLOHEX-4-ENE-1,2,3-TRIOL, PERIPLASMIC TREHALASE | | Authors: | Gibson, R.P, Gloster, T.M, Roberts, S, Warren, R.A.J, Storch De Gracia, I, Garcia, A, Chiara, J.L, Davies, G.J. | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Basis for Trehalase Inhibition Revealed by the Structure of Trehalase in Complex with Potent Inhibitors.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
