2WW5
 
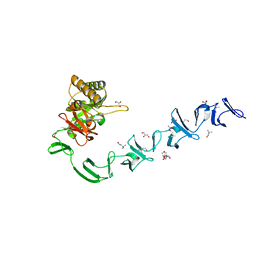 | | 3D-structure of the modular autolysin LytC from Streptococcus pneumoniae at 1.6 A resolution | | Descriptor: | 1,4-BETA-N-ACETYLMURAMIDASE, CHLORIDE ION, CHOLINE ION, ... | | Authors: | Perez-Dorado, I, Sanles, R, Hermoso, J.A, Gonzalez, A, Garcia, A, Garcia, P, Garcia, J.L, Menendez, M. | | Deposit date: | 2009-10-21 | | Release date: | 2010-04-21 | | Last modified: | 2011-09-21 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Insights Into Pneumococcal Fratricide from the Crystal Structures of the Modular Killing Factor Lytc.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2WWC
 
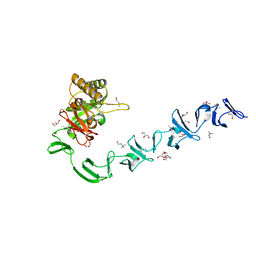 | | 3D-structure of the modular autolysin LytC from Streptococcus pneumoniae in complex with synthetic peptidoglycan ligand | | Descriptor: | 1,4-BETA-N-ACETYLMURAMIDASE, CHOLINE ION, GLYCEROL | | Authors: | Perez-Dorado, I, Sanles, R, Hermoso, J.A, Gonzalez, A, Garcia, A, Garcia, P, Garcia, J.L. | | Deposit date: | 2009-10-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights Into Pneumococcal Fratricide from the Crystal Structures of the Modular Killing Factor Lytc.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2WWD
 
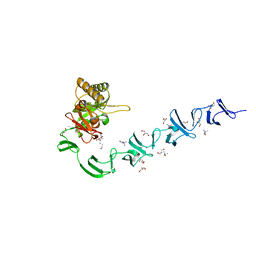 | | 3D-structure of the modular autolysin LytC from Streptococcus pneumoniae in complex with pneummococcal peptidoglycan fragment | | Descriptor: | 1,4-BETA-N-ACETYLMURAMIDASE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, ALANINE, ... | | Authors: | Perez-Dorado, I, Sanles, R, Hermoso, J.A, Gonzalez, A, Garcia, A, Garcia, P, Garcia, J.L. | | Deposit date: | 2009-10-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insights Into Pneumococcal Fratricide from the Crystal Structures of the Modular Killing Factor Lytc.
Nat.Struct.Mol.Biol., 17, 2010
|
|
8UZD
 
 | |
7YX8
 
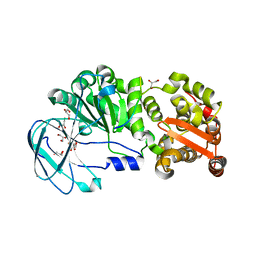 | | Crystal structure of the AM0627 (E326A) inactive mutant in complex with PSGL-1-like bis-T glycopeptide and Zn2+ | | Descriptor: | GLYCEROL, PSGL-1-like bis-T glycopeptide, Peptidase M60 domain-containing protein, ... | | Authors: | Taleb, V, Liao, Q, Narimatsu, Y, Garcia-Garcia, A, Companon, I, Borges, R.J, Gonzalez-Ramirez, A.M, Corzana, F, Clausen, H, Rovira, C, Hurtado-Guerrero, R. | | Deposit date: | 2022-02-15 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic insights into the cleavage of clustered O-glycan patches-containing glycoproteins by mucinases of the human gut.
Nat Commun, 13, 2022
|
|
6TKV
 
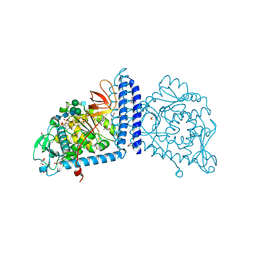 | | Crystal structure of the human FUT8 in complex with GDP and a biantennary complex N-glycan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-(1,6)-fucosyltransferase, GLYCEROL, ... | | Authors: | Garcia-Garcia, A, Ceballos-Laita, L, Serna, L, Artschwager, R, Reichardt, N.C, Corzana, F, Hurtado-Guerrero, R. | | Deposit date: | 2019-11-29 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for substrate specificity and catalysis of alpha 1,6-fucosyltransferase.
Nat Commun, 11, 2020
|
|
5NNN
 
 | |
7U4E
 
 | |
7U4F
 
 | |
8SAP
 
 | |
8SAM
 
 | |
7U4G
 
 | | Neuraminidase from influenza virus A/Shandong/9/1993(H3N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase, ... | | Authors: | Lei, R, Hernandez Garcia, A. | | Deposit date: | 2022-02-28 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Prevalence and mechanisms of evolutionary contingency in human influenza H3N2 neuraminidase.
Nat Commun, 13, 2022
|
|
8SAO
 
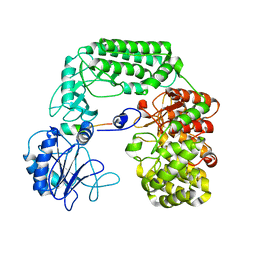 | |
5G1N
 
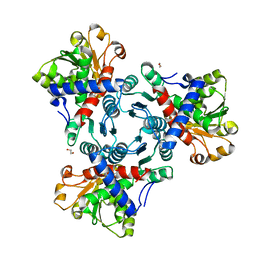 | | Aspartate transcarbamoylase domain of human CAD bound to PALA | | Descriptor: | 1,2-ETHANEDIOL, CAD PROTEIN, N-(PHOSPHONACETYL)-L-ASPARTIC ACID | | Authors: | Ruiz-Ramos, A, Grande-Garcia, A, Moreno-Morcillo, M.D, Ramon-Maiques, S. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Functional Characterization of Human Aspartate Transcarbamoylase, the Target of the Anti-Tumoral Drug Pala.
Structure, 24, 2016
|
|
5G1O
 
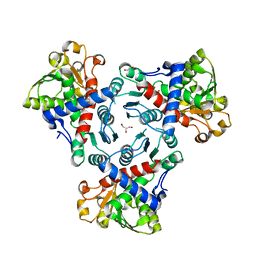 | | Aspartate transcarbamoylase domain of human CAD in apo form | | Descriptor: | 1,2-ETHANEDIOL, CAD protein, GLYCEROL | | Authors: | Ruiz-Ramos, A, Grande-Garcia, A, Moreno-Morcillo, M.D, Ramon-Maiques, S. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Functional Characterization of Human Aspartate Transcarbamoylase, the Target of the Anti-Tumoral Drug Pala.
Structure, 24, 2016
|
|
5G1P
 
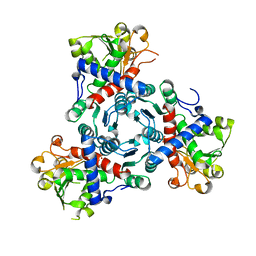 | | Aspartate transcarbamoylase domain of human CAD bound to carbamoyl phosphate | | Descriptor: | CAD PROTEIN, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | Authors: | Ruiz-Ramos, A, Grande-Garcia, A, Moreno-Morcillo, M.D, Ramon-Maiques, S. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure and Functional Characterization of Human Aspartate Transcarbamoylase, the Target of the Anti-Tumoral Drug Pala.
Structure, 24, 2016
|
|
8DWB
 
 | |
5NNQ
 
 | |
7SO8
 
 | |
6HFK
 
 | |
6HFL
 
 | |
6HFE
 
 | |
6HFU
 
 | |
6HFF
 
 | |
6HFQ
 
 | |
