5ZA0
 
 | |
3AGC
 
 | | F218V mutant of the substrate-bound red chlorophyll catabolite reductase from Arabidopsis thaliana | | Descriptor: | 3-{(2Z,3S,4S)-5-[(Z)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-2-[(5R)-2-[(3-ethyl-5-formyl-4-methyl-1H-pyrrol-2-yl)methyl]-5-(methoxycarbonyl)-3-methyl-4-oxo-4,5-dihydrocyclopenta[b]pyrrol-6(1H)-ylidene]-4-methyl-3,4-dihydro-2H-pyrrol-3-yl}propanoic acid, Red chlorophyll catabolite reductase, chloroplastic, ... | | Authors: | Sugishima, M, Fukuyama, K. | | Deposit date: | 2010-03-30 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the substrate-bound forms of red chlorophyll catabolite reductase: implications for site-specific and stereospecific reaction
J.Mol.Biol., 402, 2010
|
|
2PDB
 
 | | Human aldose reductase mutant F121P complexed with zopolrestat. | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2007-03-31 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features.
J.Mol.Biol., 379, 2008
|
|
1T02
 
 | | Crystal structure of a Statin bound to class II HMG-CoA reductase | | Descriptor: | (3R,5R)-7-((1R,2R,6S,8R,8AS)-2,6-DIMETHYL-8-{[(2R)-2-METHYLBUTANOYL]OXY}-1,2,6,7,8,8A-HEXAHYDRONAPHTHALEN-1-YL)-3,5-DIHYDROXYHEPTANOIC ACID, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, SULFATE ION | | Authors: | Tabernero, L, Rodwell, V.W, Stauffacher, C. | | Deposit date: | 2004-04-07 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a statin bound to a class II hydroxymethylglutaryl-CoA reductase.
J.Biol.Chem., 278, 2003
|
|
7KSQ
 
 | | The Structure of the moss PSI-LHCI reveals the evolution of the LHCI antenna | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Riddle, R, Gorski, C, Toporik, H, Dobson, Z, Da, Z, Williams, D, Mazor, Y. | | Deposit date: | 2020-11-23 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The structure of the Physcomitrium patens photosystem I reveals a unique Lhca2 paralogue replacing Lhca4.
Nat.Plants, 8, 2022
|
|
7KUX
 
 | | The Structure of the moss PSI-LHCI reveals the evolution of the LHCI antenna | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Riddle, R, Gorski, C, Toporik, H, Dobson, Z, Da, Z, Williams, D, Mazor, Y. | | Deposit date: | 2020-11-25 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The structure of the Physcomitrium patens photosystem I reveals a unique Lhca2 paralogue replacing Lhca4.
Nat.Plants, 8, 2022
|
|
5ZIC
 
 | | Crystal structure of human GnT-V luminal domain in complex with acceptor sugar | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-6-thio-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2018-03-14 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of cancer-associated N-acetylglucosaminyltransferase-V.
Nat Commun, 9, 2018
|
|
7M8I
 
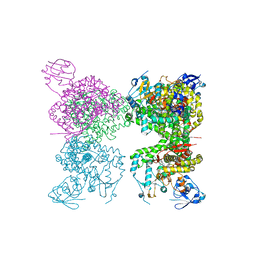 | | Human CYP11B2 and human adrenodoxin in complex with fadrozole | | Descriptor: | 4-[(5R)-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-5-yl]benzonitrile, Adrenodoxin, Cytochrome P450 11B2, ... | | Authors: | Scott, E.E, Brixius-Anderko, S. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structural and functional insights into aldosterone synthase interaction with its redox partner protein adrenodoxin.
J.Biol.Chem., 296, 2021
|
|
1OVD
 
 | | THE K136E MUTANT OF LACTOCOCCUS LACTIS DIHYDROOROTATE DEHYDROGENASE A IN COMPLEX WITH OROTATE | | Descriptor: | DIHYDROOROTATE DEHYDROGENASE A, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2003-03-26 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function.
J.Biol.Chem., 278, 2003
|
|
1JPU
 
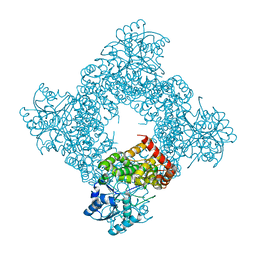 | | Crystal Structure of Bacillus Stearothermophilus Glycerol Dehydrogenase | | Descriptor: | ZINC ION, glycerol dehydrogenase | | Authors: | Ruzheinikov, S.N, Burke, J, Sedelnikova, S, Baker, P.J, Taylor, R, Bullough, P.A, Muir, N.M, Gore, M.G, Rice, D.W. | | Deposit date: | 2001-08-03 | | Release date: | 2001-10-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glycerol dehydrogenase. structure, specificity, and mechanism of a family III polyol dehydrogenase.
Structure, 9, 2001
|
|
1P5Z
 
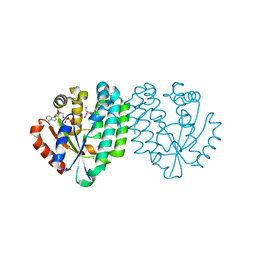 | | Structure of human dCK complexed with cytarabine and ADP-MG | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CYTARABINE, Deoxycytidine kinase, ... | | Authors: | Sabini, E, Ort, S, Monnerjahn, C, Konrad, M, Lavie, A. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of human dCK suggests strategies to improve anticancer and antiviral therapy
Nat.Struct.Biol., 10, 2003
|
|
4KPJ
 
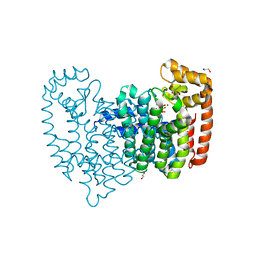 | | Crystal Structure of Farnesyl Pyrophosphate Synthase (Y204A) Mutant complexed with Mg, Pamidronate | | Descriptor: | 1,2-ETHANEDIOL, Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | Authors: | Barnett, B.L, Tsoumpra, M.K, Muniz, J.R.C, Walter, R.L. | | Deposit date: | 2013-05-13 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Farnesyl Pyrophosphate Synthase (Y204A) Mutant complexed with Mg, Pamidronate
To be Published
|
|
3L66
 
 | | Xenobiotic Reductase A - C25A Variant with Coumarin | | Descriptor: | (R,R)-2,3-BUTANEDIOL, COUMARIN, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Spiegelhauer, O, Dobbek, H. | | Deposit date: | 2009-12-23 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Cysteine as a modulator residue in the active site of xenobiotic reductase A: a structural, thermodynamic and kinetic study
J.Mol.Biol., 398, 2010
|
|
2QHX
 
 | | Structure of Pteridine Reductase from Leishmania major complexed with a ligand | | Descriptor: | IODIDE ION, METHYL 1-(4-{[(2,4-DIAMINOPTERIDIN-6-YL)METHYL](METHYL)AMINO}BENZOYL)PIPERIDINE-4-CARBOXYLATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Gibellini, F, Mcluskey, K, Tulloch, L, Hunter, W.N. | | Deposit date: | 2007-07-03 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery of potent pteridine reductase inhibitors to guide antiparasite drug development.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3AEE
 
 | | Crystal structure of porcine heart mitochondrial complex II bound with Atpenin A5 | | Descriptor: | 3-[(2S,4S,5R)-5,6-DICHLORO-2,4-DIMETHYL-1-OXOHEXYL]-4-HYDROXY-5,6-DIMETHOXY-2(1H)-PYRIDINONE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Sasaki, T, Shindo, M, Kido, Y, Inaoka, D.K, Omori, J, Osanai, A, Sakamoto, K, Mao, J, Matsuoka, S, Inoue, M, Honma, T, Tanaka, A, Kita, K. | | Deposit date: | 2010-02-04 | | Release date: | 2011-02-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Crystal structure of porcine heart mitochondrial complex II bound with Atpenin A5
To be Published
|
|
1PZS
 
 | |
3CAQ
 
 | | Crystal structure of 5beta-reductase (AKR1D1) in complex with NADPH | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3-oxo-5-beta-steroid 4-dehydrogenase, ... | | Authors: | Faucher, F, Cantin, L, Breton, R. | | Deposit date: | 2008-02-20 | | Release date: | 2008-12-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Human Delta4-3-Ketosteroid 5beta-Reductase (AKR1D1) Reveal the Presence of an Alternative Binding Site Responsible for Substrate Inhibition (dagger) (,) (double dagger).
Biochemistry, 47, 2008
|
|
3DEY
 
 | |
7L1P
 
 | | HIV Integrase Core domain (IN) in complex with dimer-spanning ligand | | Descriptor: | (2-{[3-(4-{2-[(3-{[3-(carboxymethyl)-5-methyl-1-benzofuran-2-yl]ethynyl}benzene-1-carbonyl)amino]ethyl}piperazine-1-carbonyl)phenyl]ethynyl}-5-methyl-1-benzofuran-3-yl)acetic acid, IODIDE ION, Integrase, ... | | Authors: | Gorman, M.A, Parker, M.W. | | Deposit date: | 2020-12-15 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | HIV Integrase core domain in complex with inhibitor
To Be Published
|
|
2HI7
 
 | | Crystal structure of DsbA-DsbB-ubiquinone complex | | Descriptor: | Disulfide bond formation protein B, Thiol:disulfide interchange protein dsbA, UBIQUINONE-1, ... | | Authors: | Inaba, K, Murakami, S, Suzuki, M, Nakagawa, A, Yamashita, E, Okada, K, Ito, K. | | Deposit date: | 2006-06-29 | | Release date: | 2006-12-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of the DsbB-DsbA Complex Reveals a Mechanism of Disulfide Bond Generation
Cell(Cambridge,Mass.), 127, 2006
|
|
4D78
 
 | | Cytochrome P450 3A4 bound to an inhibitor | | Descriptor: | CYTOCHROME P450 3A4, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Sevrioukova, I, Poulos, T. | | Deposit date: | 2014-11-21 | | Release date: | 2015-09-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Inhibitor Design for Evaluation of a Cyp3A4 Pharmacophore Model.
J.Med.Chem., 59, 2016
|
|
1RX1
 
 | |
1JUB
 
 | | The K136E mutant of lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-24 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
3BLX
 
 | | Yeast Isocitrate Dehydrogenase (Apo Form) | | Descriptor: | Isocitrate dehydrogenase [NAD] subunit 1, Isocitrate dehydrogenase [NAD] subunit 2 | | Authors: | Taylor, A.B, Hu, G, Hart, P.J, McAlister-Henn, L. | | Deposit date: | 2007-12-11 | | Release date: | 2008-02-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allosteric Motions in Structures of Yeast NAD+-specific Isocitrate Dehydrogenase.
J.Biol.Chem., 283, 2008
|
|
4GBU
 
 | |
