9FBL
 
 | | STRUCTURE OF HUMAN PROTEIN KINASE CK2 CATALYTIC SUBUNIT (CK2ALPHA, CSNK2A1 gene product) IN COMPLEX WITH THE CYCLIC PEPTIDOMIMETIC COMPOUND FMP35 DISCOVERED BY HIGH-THROUGHPUT SCREENING | | Descriptor: | Casein kinase II subunit alpha, Cyclic peptidomimetic compound FMP35, NICOTINIC ACID, ... | | Authors: | Werner, C, Niefind, K, Eimermacher, S, Pietsch, M, Lindenblatt, D. | | Deposit date: | 2024-05-14 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure of human protein kinase CK2 catalytic subunit (CK2alpha) in complex with the cyclic peptidomimetic compound FMP35 discovered by high-throughput screening
To Be Published
|
|
3SEP
 
 | | E. coli (lacZ) beta-galactosidase (S796A) | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-06-10 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
3S6L
 
 | |
4D65
 
 | |
2H88
 
 | | Avian Mitochondrial Respiratory Complex II at 1.8 Angstrom Resolution | | Descriptor: | AZIDE ION, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Huang, L.S, Shen, J.T, Wang, A.C, Berry, E.A. | | Deposit date: | 2006-06-06 | | Release date: | 2006-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystallographic studies of the binding of ligands to the dicarboxylate site of Complex II, and the identity of the ligand in the
Biochim.Biophys.Acta, 1757
|
|
8EK5
 
 | | Engineered scFv 10LH bound to PHOX2B/HLA-A24:02 | | Descriptor: | 10LH single chain fragment variable (scFv), Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Garfinkle, S.E, Florio, T.J, Sgourakis, N.G. | | Deposit date: | 2022-09-20 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural principles of peptide-centric chimeric antigen receptor recognition guide therapeutic expansion.
Sci Immunol, 8, 2023
|
|
4JX5
 
 | |
4JX4
 
 | |
4JX6
 
 | |
6Q6G
 
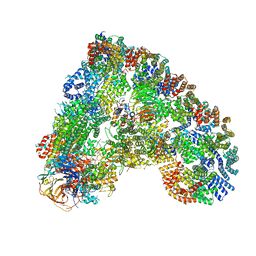 | | Cryo-EM structure of the APC/C-Cdc20-Cdk2-cyclinA2-Cks2 complex, the D1 box class | | Descriptor: | Anaphase-promoting complex subunit 1,Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Zhang, S, Barford, D. | | Deposit date: | 2018-12-11 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cyclin A2 degradation during the spindle assembly checkpoint requires multiple binding modes to the APC/C.
Nat Commun, 10, 2019
|
|
6Q6H
 
 | |
1X7K
 
 | |
9N9S
 
 | | Model of APC/C-CDC20-UBE2C from H3/H4-bound complex | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Skrajna, A, Bodrug, T, Brown, N.G, McGinty, R.K. | | Deposit date: | 2025-02-11 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | APC/C-mediated ubiquitylation of extranucleosomal histone complexes lacking canonical degrons.
Nat Commun, 16, 2025
|
|
9MJN
 
 | | Near complete virion structure of bacteriophage PhiTE | | Descriptor: | Cyanophage baseplate Pam3 plug gp18 domain-containing protein, Dit-like phage tail protein N-terminal domain-containing protein, Head stabilization/decoration protein, ... | | Authors: | Hodgkinson-Bean, J, Ayala, R. | | Deposit date: | 2024-12-16 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (12.7 Å) | | Cite: | Global structural survey of the flagellotropic myophage phi TE infecting agricultural pathogen Pectobacterium atrosepticum.
Nat Commun, 16, 2025
|
|
9N9R
 
 | | Model of APC/C-CDC20-UBE2C from H2A/H2B-bound complex | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Skrajna, A, Bodrug, T, Brown, N.G, McGinty, R.K. | | Deposit date: | 2025-02-11 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | APC/C-mediated ubiquitylation of extranucleosomal histone complexes lacking canonical degrons.
Nat Commun, 16, 2025
|
|
5MQ7
 
 | | Structure of AaLS-13 | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase | | Authors: | Sasaki, E, Boehringer, D, Leibundgut, M, Ban, N, Hilvert, D. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structure and assembly of scalable porous protein cages.
Nat Commun, 8, 2017
|
|
5A31
 
 | | Structure of the human APC-Cdh1-Hsl1-UbcH10 complex. | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Chang, L, Zhang, Z, Yang, J, Mclaughlin, S.H, Barford, D. | | Deposit date: | 2015-05-26 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Atomic Structure of the Apc/C and its Mechanism of Protein Ubiquitination.
Nature, 522, 2015
|
|
7P83
 
 | | Crystal structure of Apo form of S-adenosylmethionine synthetase from Methanocaldococcus jannaschii | | Descriptor: | S-adenosylmethionine synthase | | Authors: | Herrmann, E, Peters, A, Cornelissen, N.V, Rentmeister, A, Kuemmel, D. | | Deposit date: | 2021-07-21 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.218 Å) | | Cite: | Visible-Light Removable Photocaging Groups Accepted by MjMAT Variant: Structural Basis and Compatibility with DNA and RNA Methyltransferases.
Chembiochem, 23, 2022
|
|
1CS0
 
 | | Crystal structure of carbamoyl phosphate synthetase complexed at CYS269 in the small subunit with the tetrahedral mimic l-glutamate gamma-semialdehyde | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMOYL PHOSPHATE SYNTHETASE: LARGE SUBUNIT, CARBAMOYL PHOSPHATE SYNTHETASE: SMALL SUBUNIT, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 1999-08-16 | | Release date: | 1999-12-10 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The small subunit of carbamoyl phosphate synthetase: snapshots along the reaction pathway.
Biochemistry, 38, 1999
|
|
4UI9
 
 | | Atomic structure of the human Anaphase-Promoting Complex | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Chang, L, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2015-03-27 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Atomic Structure of the Apc and its Mechanism of Protein Ubiquitination
Nature, 522, 2015
|
|
8Z20
 
 | | Crystal structure analysis of thermotolerant Oscillatoria Phycocyanin | | Descriptor: | GLYCEROL, PHYCOCYANOBILIN, Phycocyanin subunit alpha, ... | | Authors: | Patel, S.N, Sonani, R.R, Gupta, G.D, Upadhyaya, C.T, Sonavane, B.P, Singh, N.K, Kumar, V, Madamwar, D. | | Deposit date: | 2024-04-12 | | Release date: | 2025-04-16 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and stability of phycocyanin from thermotolerant Oscillatoria.
Febs Lett., 599, 2025
|
|
8YS9
 
 | |
1CE8
 
 | | CARBAMOYL PHOSPHATE SYNTHETASE FROM ESCHERICHIS COLI WITH COMPLEXED WITH THE ALLOSTERIC LIGAND IMP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, INOSINIC ACID, ... | | Authors: | Thoden, J.B, Raushel, F.M, Holden, H.M. | | Deposit date: | 1999-03-18 | | Release date: | 1999-07-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The binding of inosine monophosphate to Escherichia coli carbamoyl phosphate synthetase.
J.Biol.Chem., 274, 1999
|
|
3CPW
 
 | | The structure of the antibiotic LINEZOLID bound to the large ribosomal subunit of HALOARCULA MARISMORTUI | | Descriptor: | 23S RIBOSOMAL RNA, 5'-R(*CP*CP*AP*(PHE)*(ACA))-3', 50S ribosomal protein L10E, ... | | Authors: | Ippolito, J.A, Kanyo, Z.K, Wang, D, Franceschi, F.J, Moore, P.B, Steitz, T.A, Duffy, E.M. | | Deposit date: | 2008-04-01 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Oxazolidinone Antibiotic
Linezolid Bound to the 50S Ribosomal Subunit
J.Med.Chem., 51, 2008
|
|
3CZJ
 
 | | E. COLI (lacZ) BETA-GALACTOSIDASE (N460T) IN COMPLEX WITH D-GALCTOPYRANOSYL-1-ONE | | Descriptor: | Beta-galactosidase, D-galactonolactone, DIMETHYL SULFOXIDE, ... | | Authors: | Huber, R.E, Dugdale, M.L, Fraser, M.E, Tammam, S.D. | | Deposit date: | 2008-04-29 | | Release date: | 2009-04-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Practical Considerations When Using Temperature to Obtain Rate Constants and Activation Thermodynamics of Enzymes with Two Catalytic Steps: Native and N460T-beta-Galactosidase (E. coli) as Examples.
Protein J., 28, 2009
|
|
