8BDA
 
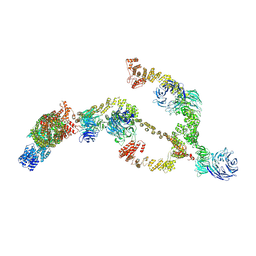 | | IFTA complex in anterograde intraflagellar transport trains (Chlamydomonas reinhardtii) | | Descriptor: | Intraflagellar transport particle protein 140, Intraflagellar transport protein 121, Intraflagellar transport protein 122 homolog, ... | | Authors: | Lacey, S.E, Foster, H.E, Pigino, G. | | Deposit date: | 2022-10-18 | | Release date: | 2023-01-11 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (20.700001 Å) | | Cite: | The molecular structure of IFT-A and IFT-B in anterograde intraflagellar transport trains.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4OWR
 
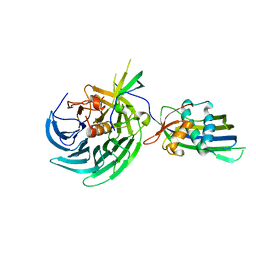 | | Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair Rae1-Nup98 | | Descriptor: | Matrix protein, Nuclear pore complex protein Nup98-Nup96, mRNA export factor | | Authors: | Ren, Y, Quan, B, Seo, H.S, Blobel, G. | | Deposit date: | 2014-02-03 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair (Rae1 Nup98).
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3MZL
 
 | | Sec13/Sec31 edge element, loop deletion mutant | | Descriptor: | Protein transport protein SEC13, Protein transport protein SEC31 | | Authors: | Whittle, J.R, Schwartz, T.U. | | Deposit date: | 2010-05-12 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Sec13-Sec16 edge element, a template for assembly of the COPII vesicle coat.
J.Cell Biol., 190, 2010
|
|
4OZU
 
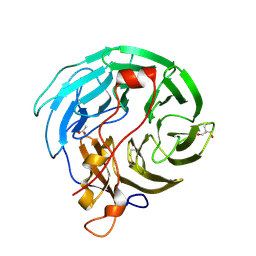 | | Crystal Structure of WD40 domain from Toxoplasma gondii coronin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Coronin | | Authors: | Kallio, J.P, Kursula, I. | | Deposit date: | 2014-02-19 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Toxoplasma gondii coronin, an actin-binding protein that relocalizes to the posterior pole of invasive parasites and contributes to invasion and egress.
Faseb J., 28, 2014
|
|
4PBY
 
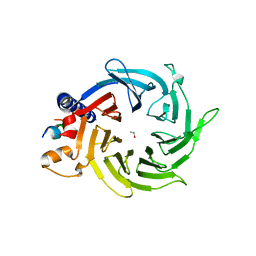 | | Structure of the human RbAp48-MTA1(656-686) complex | | Descriptor: | Histone-binding protein RBBP4, ISOPROPYL ALCOHOL, Metastasis-associated protein MTA1 | | Authors: | Murthy, A, Lejon, S, Alqarni, S.S.M, Silva, A.P.G, Watson, A.A, Mackay, J.P, Laue, E.D. | | Deposit date: | 2014-04-14 | | Release date: | 2014-06-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into the architecture of the NuRD complex: Structure of the RbAp48-MTA1 sub-complex.
J.Biol.Chem., 289, 2014
|
|
4L9O
 
 | | Crystal Structure of the Sec13-Sec16 blade-inserted complex from Pichia pastoris | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | McMahon, C, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2013-06-18 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Sec16 influences transitional ER sites by regulating rather than organizing COPII.
Mol Biol Cell, 24, 2013
|
|
4PSX
 
 | | Crystal structure of histone acetyltransferase complex | | Descriptor: | COENZYME A, Histone H3, Histone H4, ... | | Authors: | Yang, M, Li, Y. | | Deposit date: | 2014-03-08 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex
Genes Dev., 28, 2014
|
|
4PC0
 
 | | Structure of the human RbAp48-MTA1(670-711) complex | | Descriptor: | CALCIUM ION, GLYCEROL, Histone-binding protein RBBP4, ... | | Authors: | Alqarni, S.S.M, Silva, A.P.G, Mackay, J.P, Laue, E.D. | | Deposit date: | 2014-04-14 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into the architecture of the NuRD complex: Structure of the RbAp48-MTA1 sub-complex.
J.Biol.Chem., 289, 2014
|
|
4N14
 
 | | Crystal structure of Cdc20 and apcin complex | | Descriptor: | 2-(2-methyl-5-nitro-1H-imidazol-1-yl)ethyl [(1R)-2,2,2-trichloro-1-(pyrimidin-2-ylamino)ethyl]carbamate, Cell division cycle protein 20 homolog | | Authors: | Luo, X, Tian, W, Yu, H. | | Deposit date: | 2013-10-03 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C.
Nature, 514, 2014
|
|
4PBZ
 
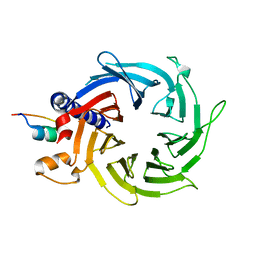 | | Structure of the human RbAp48-MTA1(670-695) complex | | Descriptor: | Histone-binding protein RBBP4, Metastasis-associated protein MTA1 | | Authors: | Murthy, A, Pei, X.Y, Watson, A.A, Silva, A.P.G, Mackay, J.P, Laue, E.D. | | Deposit date: | 2014-04-14 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Insight into the architecture of the NuRD complex: Structure of the RbAp48-MTA1 sub-complex.
J.Biol.Chem., 289, 2014
|
|
3MKS
 
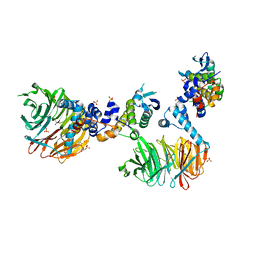 | | Crystal Structure of yeast Cdc4/Skp1 in complex with an allosteric inhibitor SCF-I2 | | Descriptor: | 1,1'-binaphthalene-2,2'-dicarboxylic acid, Cell division control protein 4, GLYCEROL, ... | | Authors: | Orlicky, S, Sicheri, F, Tyers, M, Tang, X. | | Deposit date: | 2010-04-15 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase.
Nat.Biotechnol., 28, 2010
|
|
3MKQ
 
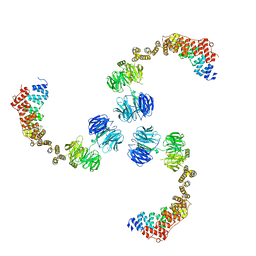 | |
4PSW
 
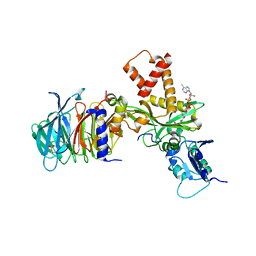 | | Crystal structure of histone acetyltransferase complex | | Descriptor: | COENZYME A, Histone H4 type VIII, Histone acetyltransferase type B catalytic subunit, ... | | Authors: | Yang, M, Li, Y. | | Deposit date: | 2014-03-08 | | Release date: | 2014-07-09 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex
Genes Dev., 28, 2014
|
|
4R7A
 
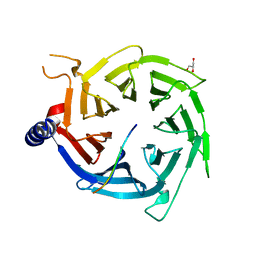 | | Crystal Structure of RBBP4 bound to PHF6 peptide | | Descriptor: | GLYCEROL, Histone-binding protein RBBP4, PHD finger protein 6 | | Authors: | Liu, Z, Li, F, Zhang, B, Li, S, Wu, J, Shi, Y. | | Deposit date: | 2014-08-27 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of Plant Homeodomain Finger 6 (PHF6) Recognition by the Retinoblastoma Binding Protein 4 (RBBP4) Component of the Nucleosome Remodeling and Deacetylase (NuRD) Complex
J.Biol.Chem., 290, 2015
|
|
6QK7
 
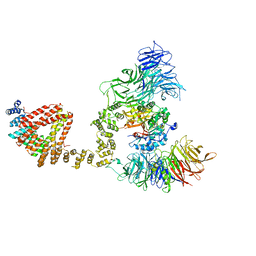 | | Elongator catalytic subcomplex Elp123 lobe | | Descriptor: | 5'-DEOXYADENOSINE, Elongator complex protein 1, Elongator complex protein 2, ... | | Authors: | Dauden, M.I, Jaciuk, M, Glatt, S. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis of tRNA recognition by the Elongator complex.
Sci Adv, 5, 2019
|
|
5WAK
 
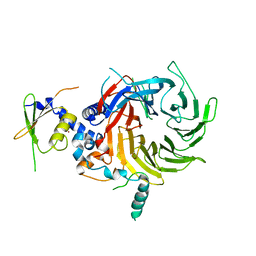 | | Crystal Structure of a Suz12-Rbbp4 Binary Complex | | Descriptor: | Histone-binding protein RBBP4, Polycomb protein SUZ12, ZINC ION | | Authors: | Chen, S, Jiao, L, Liu, X. | | Deposit date: | 2017-06-26 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Unique Structural Platforms of Suz12 Dictate Distinct Classes of PRC2 for Chromatin Binding.
Mol. Cell, 69, 2018
|
|
5WBL
 
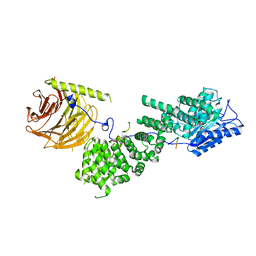 | |
5WBK
 
 | |
5WUK
 
 | |
5WJC
 
 | | Crystal structure of Schizosaccharomyces pombe Mis16 in complex with Eic1 | | Descriptor: | Eic1 protein, Kinetochore protein Mis16 | | Authors: | An, S, Cho, U.-S, Koldewey, P, Chik, J, Subramanian, L. | | Deposit date: | 2017-07-21 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Mis16 Switches Function from a Histone H4 Chaperone to a CENP-ACnp1-Specific Assembly Factor through Eic1 Interaction.
Structure, 26, 2018
|
|
5WFC
 
 | |
5WAI
 
 | | Crystal Structure of a Suz12-Rbbp4-Jarid2-Aebp2 Heterotetrameric Complex | | Descriptor: | Histone-binding protein RBBP4, Jumonji, AT-rich interactive domain 2, ... | | Authors: | Chen, S, Jiao, L, Liu, X. | | Deposit date: | 2017-06-26 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Unique Structural Platforms of Suz12 Dictate Distinct Classes of PRC2 for Chromatin Binding.
Mol. Cell, 69, 2018
|
|
5WBI
 
 | |
5WF7
 
 | | Chaetomium thermophilum Polycomb Repressive Complex 2 bound to GSK126 | | Descriptor: | 1-[(2S)-butan-2-yl]-N-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3-methyl-6-[6-(piperazin-1-yl)pyridin-3-yl]-1H-indole-4-carboxamide, Histone-lysine-N-methyltransferase EZH2, Polycomb protein SUZ12 chimera, ... | | Authors: | Bratkowski, M.A, Liu, X. | | Deposit date: | 2017-07-11 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Evolutionarily Conserved Structural Platform for PRC2 Inhibition by a Class of Ezh2 Inhibitors.
Sci Rep, 8, 2018
|
|
2I3T
 
 | | Bub3 complex with Mad3 (BubR1) GLEBS motif | | Descriptor: | Cell cycle arrest protein, Spindle assembly checkpoint component | | Authors: | Larsen, N.A, Harrison, S.C. | | Deposit date: | 2006-08-20 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of Bub3 interactions in the mitotic spindle checkpoint.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
