6L69
 
 | | Crystal structure of CYP154C2 from Streptomyces avermitilis | | Descriptor: | Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, L.H, Fushinobu, S. | | Deposit date: | 2019-10-28 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Regio- and stereoselective hydroxylation of testosterone by a novel cytochrome P450 154C2 from Streptomyces avermitilis.
Biochem.Biophys.Res.Commun., 522, 2020
|
|
4LOT
 
 | | C1s CUB2-CCP1-CCP2 | | Descriptor: | Complement C1s subcomponent heavy chain | | Authors: | Wallis, R, Venkatraman Girija, U, Moody, P.C.E, Marshall, J.E. | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4DHF
 
 | | Structure of Aurora A mutant bound to Biogenidec cpd 15 | | Descriptor: | 7-cyclopentyl-2-({1-methyl-5-[(4-methylpiperazin-1-yl)carbonyl]-1H-pyrrol-3-yl}amino)-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Aurora kinase A, MAGNESIUM ION, ... | | Authors: | Silvian, L, Marcotte, D.J. | | Deposit date: | 2012-01-27 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of 2,6,7-trisubstituted-7H-pyrrolo[2,3-d]pyrimidines as Aurora kinases inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
7LNR
 
 | | Structure of the avibactam-CDD-1 120 minute complex in imidazole and MPD | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K. | | Deposit date: | 2021-02-08 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
4J89
 
 | | Different photochemical events of a genetically encoded aryl azide define and modulate GFP fluorescence | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Green fluorescent protein, ... | | Authors: | Reddington, S.C, Jones, D.D, Rizkallah, P.J, Tippmann, E.M. | | Deposit date: | 2013-02-14 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Different Photochemical Events of a Genetically Encoded Phenyl Azide Define and Modulate GFP Fluorescence.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4J9O
 
 | | Human DNA polymerase eta-DNA ternary complex: primer extension after a T:G mispair | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*T*AP*CP*GP*TP*CP*AP*TP*G)-3'), ... | | Authors: | Zhao, Y, Gregory, M, Biertumpfel, C, Hua, Y, Hanaoka, F, Yang, W. | | Deposit date: | 2013-02-16 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Mechanism of somatic hypermutation at the WA motif by human DNA polymerase eta.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4J60
 
 | |
3MOK
 
 | | Structure of Apo HasAp from Pseudomonas aeruginosa to 1.55A Resolution | | Descriptor: | Heme acquisition protein HasAp, PHOSPHATE ION, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Jepkorir, G, Rodriguez, J.C, Rui, H, Im, W, Alontaga, A.Y, Yukl, E, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2010-04-22 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural, NMR Spectroscopic, and Computational Investigation of Hemin Loading in the Hemophore HasAp from Pseudomonas aeruginosa.
J.Am.Chem.Soc., 132, 2010
|
|
3KLL
 
 | | Crystal structure of Lactobacillus reuteri N-terminally truncated glucansucrase GTF180-maltose complex | | Descriptor: | CALCIUM ION, GLYCEROL, Glucansucrase, ... | | Authors: | Vujicic-Zagar, A, Pijning, T, Kralj, S, Eeuwema, W, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2009-11-08 | | Release date: | 2010-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3M5Z
 
 | | Crystal structure of the mutant V182A,I218A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum | | Descriptor: | Orotidine 5'-phosphate decarboxylase, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-03-14 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
4J64
 
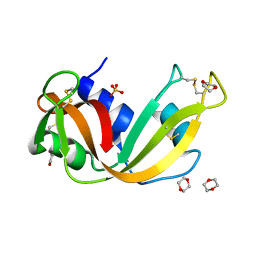 | | Crystal structure of Ribonuclease A soaked in 40% Dioxane: One of twelve in MSCS set | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kearney, B.M, Dechene, M, Swartz, P.D, Mattos, C. | | Deposit date: | 2013-02-11 | | Release date: | 2014-01-22 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | DRoP: A program for analysis of water structure on protein surfaces
to be published
|
|
4DNM
 
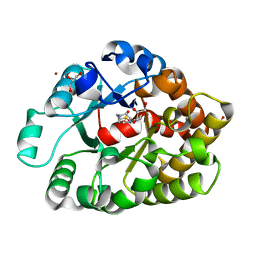 | | Crystal structure of an amidohydrolase (cog3618) from burkholderia multivorans (target efi-500235) with bound hepes, space group p3221 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Amidohydrolase 2, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N.F, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-08 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of an amidohydrolase (cog3618) from burkholderia multivorans (target efi-500235) with bound hepes, space group p3221
to be published
|
|
4DNX
 
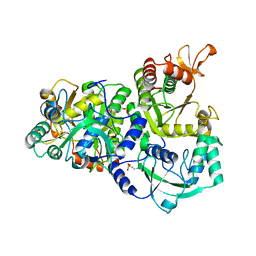 | | The structure of the ATP sulfurylase from Allochromatium vinosum in the open state | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Sulfate adenylyltransferase | | Authors: | Parey, K, Demmer, U, Warkentin, E, Dahl, C, Ermler, U. | | Deposit date: | 2012-02-09 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, biochemical and genetic characterization of dissimilatory ATP sulfurylase from Allochromatium vinosum.
Plos One, 8, 2013
|
|
4DF1
 
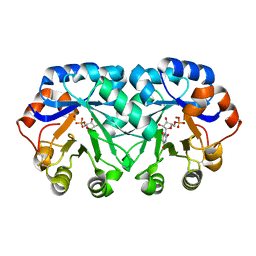 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Thermoproteus neutrophilus complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, NICKEL (II) ION, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-01-22 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from Thermoproteus neutrophilus complexed with inhibitor BMP
To be Published
|
|
3M6P
 
 | |
4JE7
 
 | | Crystal structure of a human-like mitochondrial peptide deformylase in complex with actinonin | | Descriptor: | ACTINONIN, Peptide deformylase 1A, chloroplastic/mitochondrial, ... | | Authors: | Fieulaine, S, Meinnel, T, Giglione, C. | | Deposit date: | 2013-02-26 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Understanding the highly efficient catalysis of prokaryotic peptide deformylases by shedding light on the determinants specifying the low activity of the human counterpart.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7LNO
 
 | | Structure of apo-CDD-1 beta-lactamase in imidazole and MPD | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, SULFATE ION | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K. | | Deposit date: | 2021-02-08 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | In Crystallo Time-Resolved Interaction of the Clostridioides difficile CDD-1 enzyme with Avibactam Provides New Insights into the Catalytic Mechanism of Class D beta-lactamases.
Acs Infect Dis., 7, 2021
|
|
4JAA
 
 | | Factor inhibiting HIF-1 alpha in complex with consensus ankyrin repeat domain-(d)LEU peptide | | Descriptor: | CONSENSUS ANKYRIN REPEAT DOMAIN-(d)LEU, Hypoxia-inducible factor 1-alpha inhibitor, N-OXALYLGLYCINE, ... | | Authors: | Scotti, J.S, Ge, W, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-02-18 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of an oxygenase in complex with substrate
To be Published
|
|
3HAG
 
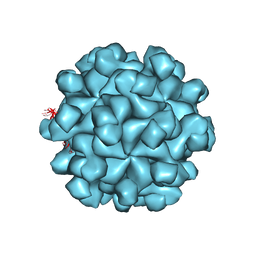 | | Crystal structure of the Hepatitis E Virus-like Particle | | Descriptor: | Capsid protein | | Authors: | Guu, T.S.Y, Liu, Z, Ye, Q, Mata, D.A, Li, K, Yin, C, Zhang, J, Tao, Y.J. | | Deposit date: | 2009-05-01 | | Release date: | 2009-09-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7L1E
 
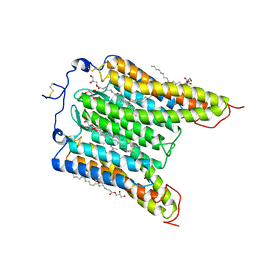 | | The Crystal Structure of Bromide-Bound GtACR1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Anion channelrhodopsin-1, BROMIDE ION, ... | | Authors: | Li, H, Huang, C.Y, Wang, M, Spudich, J.L, Zheng, L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of bromide-bound Gt ACR1 reveals a pre-activated state in the transmembrane anion tunnel.
Elife, 10, 2021
|
|
4DLH
 
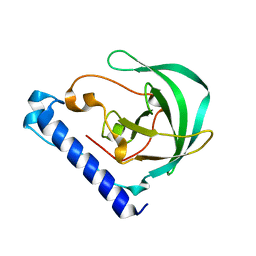 | | Crystal Structure of the protein Q9HRE7 from Halobacterium salinarium at the resolution 1.9A, Northeast Structural Genomics Consortium (NESG) Target HsR50 | | Descriptor: | uncharacterized protein | | Authors: | Kuzin, A, Chen, Y, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Northeast Structural Genomics Consortium Target HsR50
To be Published
|
|
4DUE
 
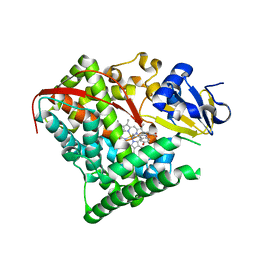 | | cytochrome P450 BM3h-2G9C6 MRI sensor bound to serotonin | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SEROTONIN, cytochrome P450 BM3 variant 2G9C6 | | Authors: | Brustad, E.M, Lelyveld, V.S, Snow, C.D, Crook, N, Martinez, F.M, Scholl, T.J, Jasanoff, A, Arnold, F.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-guided directed evolution of highly selective p450-based magnetic resonance imaging sensors for dopamine and serotonin.
J.Mol.Biol., 422, 2012
|
|
3M5X
 
 | | Crystal structure of the mutant V182A,I199A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum | | Descriptor: | Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-03-14 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
4JKU
 
 | | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with quinaldic acid, NYSGRC Target 14306 | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, Probable sugar kinase protein, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-11 | | Release date: | 2013-03-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of probable sugar kinase protein from Rhizobium etli CFN 42 complexed with quinaldic acid, NYSGRC Target 14306
To be Published
|
|
4DYB
 
 | | Crystal Structure of WSN/A Influenza Nucleoprotein with BMS-883559 Ligand Bound | | Descriptor: | N-[4-chloranyl-5-[4-[[3-(2-methoxyphenyl)-5-methyl-1,2-oxazol-4-yl]carbonyl]piperazin-1-yl]-2-nitro-phenyl]thiophene-2-carboxamide, Nucleocapsid protein | | Authors: | Lewis, H.A, Baldwin, E.T, Steinbacher, S, Maskos, K, Mortl, M, Kiefersauer, R, Edavettal, S, McDonnell, P.A, Pearce, B.C, Langley, D.R. | | Deposit date: | 2012-02-28 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | To be determined
To be Published
|
|
