3JCN
 
 | | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association: Initiation Complex I | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Sprink, T, Ramrath, D.J.F, Yamamoto, H, Yamamoto, K, Loerke, J, Ismer, J, Hildebrand, P.W, Scheerer, P, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association.
Sci Adv, 2, 2016
|
|
3K74
 
 | | Disruption of protein dynamics by an allosteric effector antibody | | Descriptor: | Dihydrofolate reductase, Nanobody | | Authors: | Oyen, D, Srinivasan, V, Steyaert, J, Barlow, J. | | Deposit date: | 2009-10-12 | | Release date: | 2010-10-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Constraining enzyme conformational change by an antibody leads to hyperbolic inhibition.
J.Mol.Biol., 407, 2011
|
|
8CTA
 
 | | Minimal 2:2 Ternary Complex between BI-224436 bound HIV-1 Integrase Catalytic Core Domain Dimer and Carboxy Terminal Domains | | Descriptor: | (2S)-tert-butoxy[4-(2,3-dihydropyrano[4,3,2-de]quinolin-7-yl)-2-methylquinolin-3-yl]acetic acid, 1,2-ETHANEDIOL, Integrase | | Authors: | Gupta, K, Van Duyne, G.D, Eilers, G, Bushman, F.D. | | Deposit date: | 2022-05-13 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structure of a HIV-1 IN-Allosteric inhibitor complex at 2.93 angstrom resolution: Routes to inhibitor optimization.
Plos Pathog., 19, 2023
|
|
5J55
 
 | | The Structure and Mechanism of NOV1, a Resveratrol-Cleaving Dioxygenase | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, Carotenoid oxygenase, FE (III) ION, ... | | Authors: | McAndrew, R.P, Pereira, J.H, Sathitsuksanoh, N, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2016-04-01 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
2FR9
 
 | | NMR structure of the alpha-conotoxin GI (SER12)-benzoylphenylalanine derivative | | Descriptor: | Alpha-conotoxin GI | | Authors: | Pashkov, V.S, Maslennikov, I.V, Kasheverov, I.E, Zhmak, M.N, Utkin, Y.N, Tsetlin, V.I, Arseniev, A.S. | | Deposit date: | 2006-01-19 | | Release date: | 2006-05-30 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Alpha-Conotoxin GI benzoylphenylalanine derivatives. (1)H-NMR structures and photoaffinity labeling of the Torpedo californica nicotinic acetylcholine receptor.
Febs J., 273, 2006
|
|
5LSO
 
 | |
6XEB
 
 | | STRUCTURE OF HUMAN HDAC2 IN COMPLEX WITH KETONE INHIBITOR (COMPOUND E) | | Descriptor: | 5-{(1S)-7,7-dihydroxy-1-[(1-methylazetidine-3-carbonyl)amino]nonyl}-2-phenyl-1H-imidazole-4-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Clausen, D. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Development of a selective HDAC inhibitor aimed at reactivating the HIV latent reservoir.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6ZL3
 
 | | CRYSTAL STRUCTURE OF HRAS IN COMPLEX WITH COMPOUND 18 and GDP | | Descriptor: | (3~{S})-3-[2-[(dimethylamino)methyl]-1~{H}-indol-3-yl]-5-oxidanyl-2,3-dihydroisoindol-1-one, GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kessler, D, Fischer, G, Boettcher, J. | | Deposit date: | 2020-06-30 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Drugging all RAS isoforms with one pocket.
Future Med Chem, 12, 2020
|
|
6R8H
 
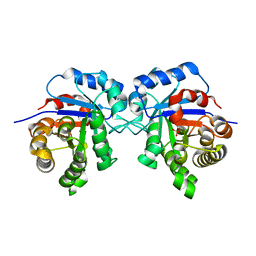 | | Triosephosphate isomerase from liver fluke (Fasciola hepatica). | | Descriptor: | SULFATE ION, Triosephosphate isomerase | | Authors: | Ferraro, F, Corvo, I, Bergalli, L, Ilarraz, A, Cabrera, M, Gil, J, Susuki, B, Caffrey, C, Timson, D.J, Robert, X, Guillon, C, Alvarez, G. | | Deposit date: | 2019-04-01 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel and selective inactivators of Triosephosphate isomerase with anti-trematode activity.
Sci Rep, 10, 2020
|
|
2UY3
 
 | |
7AWE
 
 | | HUMAN IMMUNOPROTEASOME 20S PARTICLE IN COMPLEX WITH [(1R)-2-(1-benzofuran-3-yl)-1-{[(1S,2R,4R)-7-oxabicyclo[2.2.1]heptan-2-yl]formamido}ethyl]boronic acid | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Musil, D, Klein, M. | | Deposit date: | 2020-11-06 | | Release date: | 2021-06-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | M3258 Is a Selective Inhibitor of the Immunoproteasome Subunit LMP7 ( beta 5i) Delivering Efficacy in Multiple Myeloma Models.
Mol.Cancer Ther., 20, 2021
|
|
6XGV
 
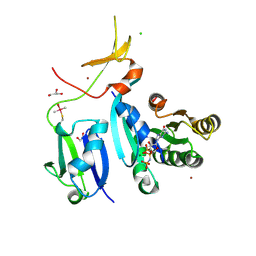 | | Crystal Structure of KRAS-G13D (GMPPNP-bound) in complex with RAS-binding domain (RBD) and cysteine-rich domain (CRD) of RAF1/CRAF | | Descriptor: | CHLORIDE ION, GLYCEROL, GTPase KRas, ... | | Authors: | Tran, T.H, Chan, A.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation.
Nat Commun, 12, 2021
|
|
1CDB
 
 | |
5JTF
 
 | | Crystal structure of ArsN N-acetyltransferase from Pseudomonas putida KT2440 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Putative Phosphinothricin N-acetyltransferase | | Authors: | Venkadesh, S, Dheeman, D.S, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2016-05-09 | | Release date: | 2017-05-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.156 Å) | | Cite: | Arsinothricin, an arsenic-containing non-proteinogenic amino acid analog of glutamate, is a broad-spectrum antibiotic.
Commun Biol, 2, 2019
|
|
2V24
 
 | | Structure of the human SPRY domain-containing SOCS box protein SSB-4 | | Descriptor: | NICKEL (II) ION, SPRY DOMAIN-CONTAINING SOCS BOX PROTEIN 4 | | Authors: | Uppenberg, J, Bullock, A, Keates, T, Savitsky, P, Pike, A.C.W, Ugochukwu, E, Bunkoczi, G, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Knapp, S. | | Deposit date: | 2007-05-31 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Par-4 Recognition by the Spry Domain-and Socs Box-Containing Proteins Spsb1, Spsb2, and Spsb4.
J.Mol.Biol., 401, 2010
|
|
5JED
 
 | | Apo-structure of humanised RadA-mutant humRadA28 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-18 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5J71
 
 | | Crystal structure of pyruvate dehydrogenase kinase isoform 2 in complex with inhibitor PS35 | | Descriptor: | 4-({5-[(piperidin-4-yl)amino]-1,3-dihydro-2H-isoindol-2-yl}sulfonyl)benzene-1,3-diol, L(+)-TARTARIC ACID, [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 2, ... | | Authors: | Gui, W.J, Tso, S.C, Chuang, J.L, Wu, C.Y, Qi, X, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2016-04-05 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Development of Dihydroxyphenyl Sulfonylisoindoline Derivatives as Liver-Targeting Pyruvate Dehydrogenase Kinase Inhibitors.
J. Med. Chem., 60, 2017
|
|
3K3K
 
 | | Crystal structure of dimeric abscisic acid (ABA) receptor pyrabactin resistance 1 (PYR1) with ABA-bound closed-lid and ABA-free open-lid subunits | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1 | | Authors: | Arvai, A.S, Hitomi, K, Getzoff, E.D. | | Deposit date: | 2009-10-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural mechanism of abscisic acid binding and signaling by dimeric PYR1.
Science, 326, 2009
|
|
9GMD
 
 | | MukEF in complex with the phage protein gp5.9 (focus) | | Descriptor: | Chromosome partition protein MukE, Chromosome partition protein MukF, Probable RecBCD inhibitor gp5.9 | | Authors: | Burmann, F, Wilkinson, O, Kimanius, D, Dillingham, M, Lowe, J. | | Deposit date: | 2024-08-28 | | Release date: | 2025-03-26 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanism of DNA capture by the MukBEF SMC complex and its inhibition by a viral DNA mimic.
Cell, 188, 2025
|
|
2V9C
 
 | | X-ray Crystallographic Structure of a Pseudomonas aeruginosa Azoreductase in Complex with Methyl Red. | | Descriptor: | 2-(4-DIMETHYLAMINOPHENYL)DIAZENYLBENZOIC ACID, FLAVIN MONONUCLEOTIDE, FMN-DEPENDENT NADH-AZOREDUCTASE 1, ... | | Authors: | Wang, C.-J, Hagemeier, C, Rahman, N, Lowe, E.D, Noble, M.E.M, Coughtrie, M, Sim, E, Westwood, I.M. | | Deposit date: | 2007-08-23 | | Release date: | 2007-11-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Molecular Cloning, Characterisation and Ligand- Bound Structure of an Azoreductase from Pseudomonas Aeruginosa
J.Mol.Biol., 373, 2007
|
|
6P7H
 
 | |
2V9J
 
 | | Crystal structure of the regulatory fragment of mammalian AMPK in complexes with Mg.ATP-AMP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Heath, R, Saiu, P, Leiper, F.C, Leone, P, Jing, C, Walker, P.A, Haire, L, Eccleston, J.F, Davis, C.T, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2007-08-23 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural Basis for AMP Binding to Mammalian AMP-Activated Protein Kinase
Nature, 449, 2007
|
|
8S6F
 
 | | A Structural Investigation of the Interaction between a GC-376-Based Peptidomimetic PROTAC and Its Precursor with the Viral Main Protease of Coxsackievirus B3 | | Descriptor: | Protease 3C, methyl (4~{S})-4-[[(2~{S})-4-methyl-2-(phenylmethoxycarbonylamino)pentanoyl]amino]-5-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | De Santis, A, Grifagni, D, Orsetti, A, Lenci, E, Rosato, A, D'Onofrio, M, Trabocchi, A, Ciofi-Baffoni, S, Cantini, F, Calderone, V. | | Deposit date: | 2024-02-27 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A Structural Investigation of the Interaction between a GC-376-Based Peptidomimetic PROTAC and Its Precursor with the Viral Main Protease of Coxsackievirus B3.
Biomolecules, 14, 2024
|
|
7QKD
 
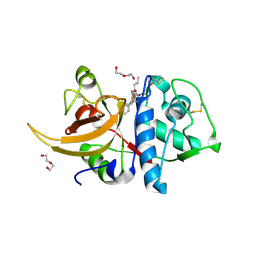 | | Crystal structure of human Cathepsin L in complex with covalently bound MG132 | | Descriptor: | ACETATE ION, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 2024
|
|
6ZIR
 
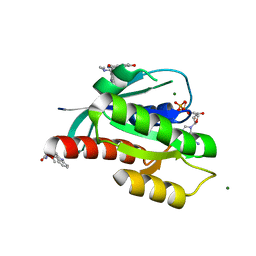 | | CRYSTAL STRUCTURE OF NRAS (C118S) IN COMPLEX WITH GDP AND COMPOUND 18 | | Descriptor: | (3~{S})-3-[2-[(dimethylamino)methyl]-1~{H}-indol-3-yl]-5-oxidanyl-2,3-dihydroisoindol-1-one, GTPase NRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kessler, D, Fischer, G, Boettcher, J. | | Deposit date: | 2020-06-26 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Drugging all RAS isoforms with one pocket.
Future Med Chem, 12, 2020
|
|
