4KCJ
 
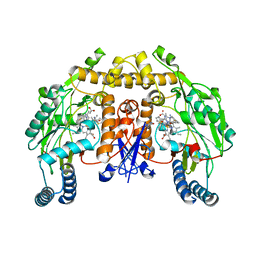 | | Structure of neuronal nitric oxide synthase heme domain in complex with N,N'-((ethane-1,2-diylbis(oxy))bis(3,1-phenylene))bis(thiophene-2-carboximidamide) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N,N'-[ethane-1,2-diylbis(oxybenzene-3,1-diyl)]dithiophene-2-carboximidamide, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Potent and Selective Double-Headed Thiophene-2-carboximidamide Inhibitors of Neuronal Nitric Oxide Synthase for the Treatment of Melanoma.
J.Med.Chem., 57, 2014
|
|
7LHB
 
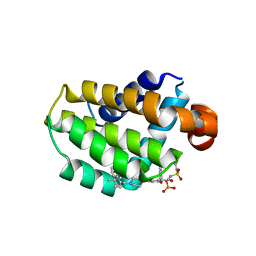 | | Crystal structure of Bcl-2 in complex with prodrug ABBV-167 | | Descriptor: | Apoptosis regulator Bcl-2, Phosphoric acid mono-[5-(5-{4-[2-(4-chloro-phenyl)-4,4-dimethyl-cyclohex-1-enylmethyl]-piperazin-1-yl}-2-{3-nitro-4-[(tetrahydro-pyran-4-ylmethyl)-amino]-benzenesulfonylaminocarbonyl}-phenoxy)-pyrrolo[2,3-b]pyridin-7-ylmethyl] ester | | Authors: | Judge, R.A, Salem, A.H. | | Deposit date: | 2021-01-21 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.068 Å) | | Cite: | Expanding the Repertoire for "Large Small Molecules": Prodrug ABBV-167 Efficiently Converts to Venetoclax with Reduced Food Effect in Healthy Volunteers.
Mol.Cancer Ther., 20, 2021
|
|
7M1V
 
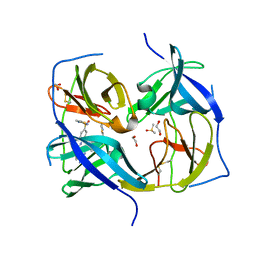 | | Structure of Zika virus NS2b-NS3 protease mutant binding the compound NSC86314 in the super-open conformation | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-{2,4-diamino-5-[2-(4-{[(2E)-1,3-thiazolidin-2-ylidene]sulfamoyl}phenyl)hydrazinyl]phenyl}hydrazinyl)-N-[(2S)-1,3-thiazolidin-2-yl]benzene-1-sulfonamide, CHLORIDE ION, ... | | Authors: | Aleshin, A.E, Shiryaev, S.A, Liddington, R.C. | | Deposit date: | 2021-03-15 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | To be provided
To Be Published
|
|
1ZFK
 
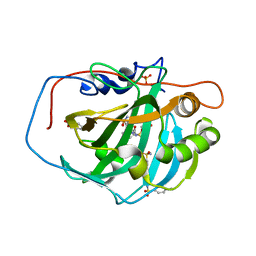 | | carbonic anhydrase II in complex with N-4-sulfonamidphenyl-N'-4-methylbenzosulfonylurease as sulfonamide inhibitor | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, Carbonic anhydrase II, GLYCEROL, ... | | Authors: | Honndorf, V.S, Heine, A, Klebe, G, Supuran, C.T. | | Deposit date: | 2005-04-20 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | carbonic anhydrase II in complex with N-4-sulfonamidphenyl-N'-4-methylbenzosulfonylurease as sulfonamide inhibitor
To be Published
|
|
2ZGA
 
 | | HIV-1 protease in complex with a dimethylallyl decorated pyrrolidine based inhibitor (hexagonal space group) | | Descriptor: | (3S,4S),-3,4-Bis-[(4-carbamoyl-benzensulfonyl)-(3-methyl-but-2-enyl)-amino]-pyrrolidine, Protease | | Authors: | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2008-01-21 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Two Solutions for the Same Problem: Multiple Binding Modes of Pyrrolidine-Based HIV-1 Protease Inhibitors
J.Mol.Biol., 410, 2011
|
|
7L9A
 
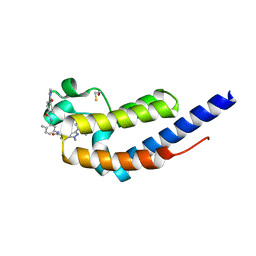 | | Crystal structure of BRDT bromodomain 2 in complex with CDD-1102 | | Descriptor: | BETA-MERCAPTOETHANOL, Bromodomain testis-specific protein, N~1~-(5-{[3-(4-amino-2-methylphenyl)-1-methyl-1H-indazole-5-carbonyl]amino}-2-methylphenyl)-N~4~-methylbenzene-1,4-dicarboxamide | | Authors: | Sharma, R, Kaur, G, Yu, Z, Ku, A.F, Anglin, J.L, Ucisik, M.N, Faver, J.C, Sankaran, B, Kim, C, Matzuk, M.M. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery and characterization of bromodomain 2-specific inhibitors of BRDT.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3AAV
 
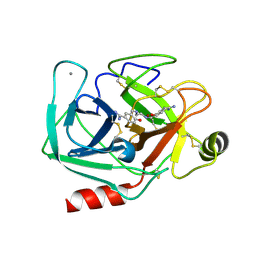 | | Bovine beta-trypsin bound to meta-diamidino schiff base copper (II) chelate | | Descriptor: | 3,3'-[ethane-1,2-diylbis(nitrilomethylylidene)]bis(4-hydroxybenzenecarboximidamide), CALCIUM ION, COPPER (II) ION, ... | | Authors: | Iyaguchi, D, Kawano, S, Toyota, E. | | Deposit date: | 2009-11-26 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the design of novel Schiff base metal chelate inhibitors of trypsin
Bioorg.Med.Chem., 18, 2010
|
|
4NI8
 
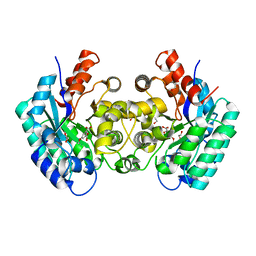 | | Crystal structure of 5-carboxyvanillate decarboxylase LigW from Sphingomonas paucimobilis complexed with Mn and 5-methoxyisophtalic acid | | Descriptor: | 1,2-ETHANEDIOL, 5-carboxyvanillate decarboxylase, 5-methoxybenzene-1,3-dicarboxylic acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Vladimirova, A, Raushel, F.M, Almo, S.C. | | Deposit date: | 2013-11-05 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of 5-carboxyvanillate decarboxylase LigW from Sphingomonas paucimobilis complexed with Mn and 5-methoxyisophtalic acid
To be Published
|
|
2PQZ
 
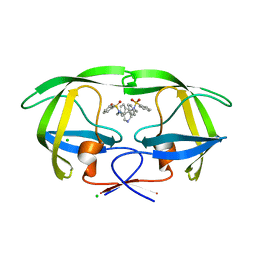 | | HIV-1 Protease in complex with a pyrrolidine-based inhibitor | | Descriptor: | CHLORIDE ION, N,N'-(3S,4S)-PYRROLIDINE-3,4-DIYLBIS(N-BENZYLBENZENESULFONAMIDE), Protease | | Authors: | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2007-05-03 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Guided Design of C2-Symmetric HIV-1 Protease Inhibitors Based on a Pyrrolidine Scaffold.
J.Med.Chem., 51, 2008
|
|
4N6D
 
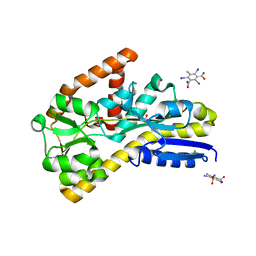 | | Crystal structure of a TRAP periplasmic solute binding protein from Desulfovibrio salexigens DSM2638 (Desal_3247), Target EFI-510112, phased with I3C, open complex, C-terminus of symmetry mate bound in ligand binding site | | Descriptor: | 1,2-ETHANEDIOL, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-11 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N87
 
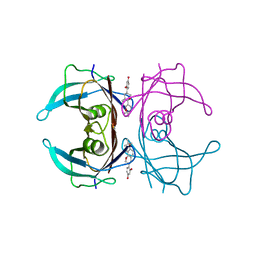 | | Crystal structure of V30M mutant human transthyretin complexed with glabridin | | Descriptor: | 4-[(3R)-8,8-dimethyl-3,4-dihydro-2H,8H-pyrano[2,3-f]chromen-3-yl]benzene-1,3-diol, Transthyretin | | Authors: | Yokoyama, T, Kosaka, Y, Mizuguchi, M. | | Deposit date: | 2013-10-17 | | Release date: | 2014-02-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Crystal structures of human transthyretin complexed with glabridin
J.Med.Chem., 57, 2014
|
|
2OS8
 
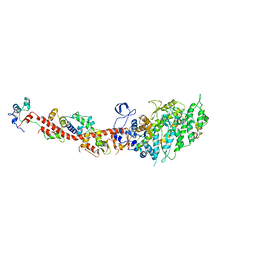 | | Rigor-like structures of muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Myosin essential light chain, ... | | Authors: | Yang, Y, Gourinath, S, Cohen, C, Brown, J.H. | | Deposit date: | 2007-02-05 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Rigor-like Structures from Muscle Myosins Reveal Key Mechanical Elements in the Transduction Pathways of This Allosteric Motor.
Structure, 15, 2007
|
|
3BYS
 
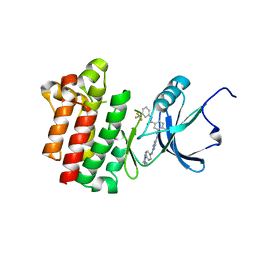 | | co-crystal structure of Lck and aminopyrimidine amide 10b | | Descriptor: | 4-methyl-N~3~-(2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-5-yl)-N~1~-[3-(trifluoromethyl)phenyl]benzene-1,3-dicarboxamide, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Huang, X. | | Deposit date: | 2008-01-16 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-guided design of aminopyrimidine amides as potent, selective inhibitors of lymphocyte specific kinase: synthesis, structure-activity relationships, and inhibition of in vivo T cell activation.
J.Med.Chem., 51, 2008
|
|
1Y91
 
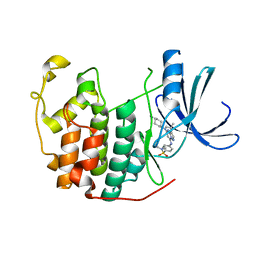 | | Crystal structure of human CDK2 complexed with a pyrazolo[1,5-a]pyrimidine inhibitor | | Descriptor: | 4-[5-(TRANS-4-AMINOCYCLOHEXYLAMINO)-3-ISOPROPYLPYRAZOLO[1,5-A]PYRIMIDIN-7-YLAMINO]-N,N-DIMETHYLBENZENESULFONAMIDE, Cell division protein kinase 2 | | Authors: | Williamson, D.S, Parratt, M.J, Torrance, C.J, Bower, J.F, Moore, J.D, Richardson, C.M, Dokurno, P, Cansfield, A.D, Francis, G.L, Hebdon, R.J, Howes, R, Jackson, P.S, Lockie, A.M, Murray, J.B, Nunns, C.L, Powles, J, Robertson, A, Surgenor, A.E. | | Deposit date: | 2004-12-14 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-guided design of pyrazolo[1,5-a]pyrimidines as inhibitors of human cyclin-dependent kinase 2.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1YGC
 
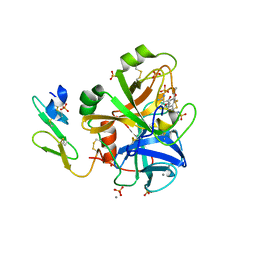 | | Short Factor VIIa with a small molecule inhibitor | | Descriptor: | (R)-4-[2-(3-AMINO-BENZENESULFONYLAMINO)-1-(3,5-DIETHOXY-2-FLUOROPHENYL)-2-OXO-ETHYLAMINO]-2-HYDROXY-BENZAMIDINE, CALCIUM ION, Coagulation factor VII, ... | | Authors: | Olivero, A.G, Eigenbrot, C, Goldsmith, R, Robarge, K, Artis, D.R, Flygare, J, Rawson, T, Refino, C, Bunting, S, Kirchhofer, D. | | Deposit date: | 2005-01-04 | | Release date: | 2005-01-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A selective, slow binding inhibitor of factor VIIa binds to a nonstandard active site conformation and attenuates thrombus formation in vivo.
J.Biol.Chem., 280, 2005
|
|
4MPE
 
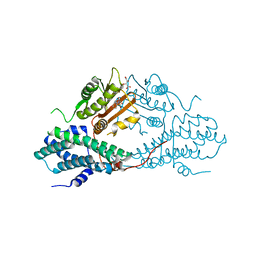 | | Crystal structure of pyruvate dehydrogenase kinase isoform 2 in complex with inhibitor PS8 | | Descriptor: | 4-[(5-hydroxy-1,3-dihydro-2H-isoindol-2-yl)sulfonyl]benzene-1,3-diol, L(+)-TARTARIC ACID, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 2, ... | | Authors: | Gui, W.J, Tso, S.C, Chuang, J.L, Wu, C.Y, Qi, X, Tambar, U.K, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2013-09-12 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Structure-guided Development of Specific Pyruvate Dehydrogenase Kinase Inhibitors Targeting the ATP-binding Pocket.
J.Biol.Chem., 289, 2014
|
|
4MPC
 
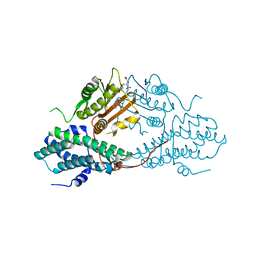 | | Crystal structure of pyruvate dehydrogenase kinase isoform 2 in complex with inhibitor PS2 | | Descriptor: | 4-(isoindolin-2-ylsulfonyl)benzene-1,3-diol, L(+)-TARTARIC ACID, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 2, ... | | Authors: | Gui, W.J, Tso, S.C, Chuang, J.L, Wu, C.Y, Qi, X, Tambar, U.K, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2013-09-12 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structure-guided Development of Specific Pyruvate Dehydrogenase Kinase Inhibitors Targeting the ATP-binding Pocket.
J.Biol.Chem., 289, 2014
|
|
252L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-10-28 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
4NSY
 
 | | Wild-type lysobacter enzymogenes lysc endoproteinase covalently inhibited by TLCK | | Descriptor: | CALCIUM ION, CHLORIDE ION, Lysyl endopeptidase, ... | | Authors: | Asztalos, P, Muller, A, Holke, W, Sobek, H, Rudolph, M.G. | | Deposit date: | 2013-11-29 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution structure of a lysine-specific endoproteinase from Lysobacter enzymogenes suggests a hydroxyl group bound to the oxyanion hole.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NC7
 
 | | N-terminal domain of delta-subunit of RNA polymerase complexed with I3C and nickel ions | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, DNA-directed RNA polymerase subunit delta, NICKEL (II) ION | | Authors: | Demo, G, Papouskova, V, Komarek, J, Sanderova, H, Rabatinova, A, Krasny, L, Zidek, L, Sklenar, V, Wimmerova, M. | | Deposit date: | 2013-10-24 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray vs. NMR structure of N-terminal domain of delta-subunit of RNA polymerase.
J.Struct.Biol., 187, 2014
|
|
3CJF
 
 | | Crystal structure of VEGFR2 in complex with a 3,4,5-trimethoxy aniline containing pyrimidine | | Descriptor: | N~4~-(3-methyl-1H-indazol-6-yl)-N~2~-(3,4,5-trimethoxyphenyl)pyrimidine-2,4-diamine, SULFATE ION, Vascular endothelial growth factor receptor 2 | | Authors: | Nolte, R.T. | | Deposit date: | 2008-03-12 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of 5-[[4-[(2,3-dimethyl-2H-indazol-6-yl)methylamino]-2-pyrimidinyl]amino]-2-methyl-benzenesulfonamide (Pazopanib), a novel and potent vascular endothelial growth factor receptor inhibitor.
J.Med.Chem., 51, 2008
|
|
4NCW
 
 | | foldon domain wild type C-conjugate | | Descriptor: | Fibritin, N,N',N''-triethylbenzene-1,3,5-tricarboxamide | | Authors: | Graewert, M.A, Berthelmann, A, Lach, J, Groll, M, Eichler, J. | | Deposit date: | 2013-10-25 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Versatile C(3)-symmetric scaffolds and their use for covalent stabilization of the foldon trimer.
Org.Biomol.Chem., 12, 2014
|
|
2R43
 
 | | I50V HIV-1 protease in complex with an amino decorated pyrrolidine-based inhibitor | | Descriptor: | CHLORIDE ION, GLYCEROL, N,N'-(3S,4S)-PYRROLIDINE-3,4-DIYLBIS(4-AMINO-N-BENZYLBENZENESULFONAMIDE), ... | | Authors: | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2007-08-30 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural and Kinetic Analysis of Pyrrolidine-Based Inhibitors of the Drug-Resistant Ile84Val Mutant of HIV-1 Protease
J.Mol.Biol., 383, 2008
|
|
2QCB
 
 | | T7-tagged full-length streptavidin complexed with ruthenium ligand | | Descriptor: | N-(4-{[(2-AMINOETHYL)AMINO]SULFONYL}PHENYL)-5-[(3AS,4S,6AR)-2-OXOHEXAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-4-YL]PENTANAMIDE-(1,2,3,4,5,6-ETA)-BENZENE-CHLORO-RUTHENIUM(III), Streptavidin | | Authors: | Le Trong, I, Creus, M, Pordea, A, Ward, T.R, Stenkamp, R.E. | | Deposit date: | 2007-06-19 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray structure and designed evolution of an artificial transfer hydrogenase
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
4NSV
 
 | | Lysobacter enzymogenes lysc endoproteinase K30R mutant covalently inhibited by TLCK | | Descriptor: | CHLORIDE ION, Lysyl endopeptidase, N-[(2S,3S)-7-amino-1-chloro-2-hydroxyheptan-3-yl]-4-methylbenzenesulfonamide (Bound Form), ... | | Authors: | Asztalos, P, Muller, A, Holke, W, Sobek, H, Rudolph, M.G. | | Deposit date: | 2013-11-29 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Atomic resolution structure of a lysine-specific endoproteinase from Lysobacter enzymogenes suggests a hydroxyl group bound to the oxyanion hole.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
