8FLM
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP, NVS-STG2 and C53 | | Descriptor: | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide, 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, ... | | Authors: | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
8FLK
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP and NVS-STG2 | | Descriptor: | 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, cGAMP | | Authors: | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
4CEV
 
 | | ARGINASE FROM BACILLUS CALDEVELOX, L-ORNITHINE COMPLEX | | Descriptor: | GUANIDINE, L-ornithine, MANGANESE (II) ION, ... | | Authors: | Bewley, M.C, Jeffrey, P.D, Patchett, M.L, Kanyo, Z.F, Baker, E.N. | | Deposit date: | 1999-03-15 | | Release date: | 1999-04-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily.
Structure Fold.Des., 7, 1999
|
|
3CEV
 
 | | ARGINASE FROM BACILLUS CALDEVELOX, COMPLEXED WITH L-ARGININE | | Descriptor: | ARGININE, MANGANESE (II) ION, PROTEIN (ARGINASE) | | Authors: | Bewley, M.C, Jeffrey, P.D, Patchett, M.L, Kanyo, Z.F, Baker, E.N. | | Deposit date: | 1999-03-15 | | Release date: | 1999-04-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily.
Structure Fold.Des., 7, 1999
|
|
8GT6
 
 | | human STING With agonist HB3089 | | Descriptor: | 1-[(2E)-4-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-[3-(morpholin-4-yl)propoxy]-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methyl-1H-furo[3,2-e]benzimidazole-5-carboxamide, Stimulator of interferon genes protein | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
8GSZ
 
 | | Structure of STING SAVI-related mutant V147L | | Descriptor: | Stimulator of interferon genes protein | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
3NJT
 
 | | Crystal structure of the R450A mutant of the membrane protein FhaC | | Descriptor: | Filamentous hemagglutinin transporter protein fhaC | | Authors: | Clantin, B, Delattre, A.S, Jacob-Dubuisson, F, Villeret, V. | | Deposit date: | 2010-06-18 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Functional importance of a conserved sequence motif in FhaC, a prototypic member of the TpsB/Omp85 superfamily.
Febs J., 277, 2010
|
|
1KKH
 
 | | Crystal Structure of the Methanococcus jannaschii Mevalonate Kinase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Mevalonate Kinase | | Authors: | Yang, D, Shipman, L.W, Roessner, C.A, Scott, A.I, Sacchettini, J.C. | | Deposit date: | 2001-12-08 | | Release date: | 2002-03-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Methanococcus jannaschii mevalonate kinase, a member of the GHMP kinase superfamily.
J.Biol.Chem., 277, 2002
|
|
4C0E
 
 | | Structure of the NOT1 superfamily homology domain from Chaetomium thermophilum | | Descriptor: | NOT1 | | Authors: | Chen, Y, Boland, A, Raisch, T, Jonas, S, Izaurralde, E, Weichenrieder, O. | | Deposit date: | 2013-08-01 | | Release date: | 2013-10-09 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Assembly of the not Module of the Human Ccr4-not Complex
Nat.Struct.Mol.Biol., 20, 2013
|
|
4TOY
 
 | |
2D4G
 
 | |
2G3T
 
 | |
1CEV
 
 | | ARGINASE FROM BACILLUS CALDOVELOX, NATIVE STRUCTURE AT PH 5.6 | | Descriptor: | MANGANESE (II) ION, PROTEIN (ARGINASE) | | Authors: | Bewley, M.C, Jeffrey, P.D, Patchett, M.L, Kanyo, Z.F, Baker, E.N. | | Deposit date: | 1999-03-12 | | Release date: | 1999-04-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily.
Structure Fold.Des., 7, 1999
|
|
2Q1M
 
 | |
2IAK
 
 | |
1FWY
 
 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINE 1-PHOSPHATE URIDYLTRANSFERASE BOUND TO UDP-GLCNAC | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, UDP-N-ACETYLGLUCOSAMINE PYROPHOSPHORYLASE, ... | | Authors: | Brown, K, Pompeo, F, Dixon, S, Mengin-Lecreulx, D, Cambillau, C, Bourne, Y. | | Deposit date: | 2000-09-25 | | Release date: | 2000-10-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily.
EMBO J., 18, 1999
|
|
5D68
 
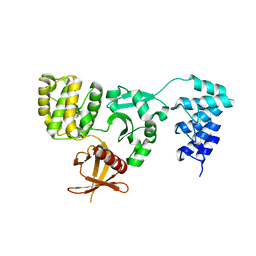 | | Crystal structure of KRIT1 ARD-FERM | | Descriptor: | Krev interaction trapped protein 1 | | Authors: | Zhang, R, Li, X, Boggon, T.J. | | Deposit date: | 2015-08-11 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Structural analysis of the KRIT1 ankyrin repeat and FERM domains reveals a conformationally stable ARD-FERM interface.
J.Struct.Biol., 192, 2015
|
|
1FXJ
 
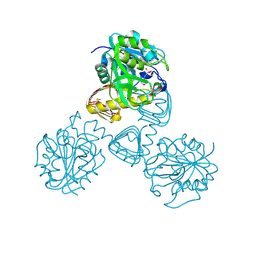 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINE 1-PHOSPHATE URIDYLTRANSFERASE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, UDP-N-ACETYLGLUCOSAMINE PYROPHOSPHORYLASE | | Authors: | Brown, K, Pompeo, F, Dixon, S, Mengin-Lecreulx, D, Cambillau, C, Bourne, Y. | | Deposit date: | 2000-09-26 | | Release date: | 2000-10-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily.
EMBO J., 18, 1999
|
|
8S9T
 
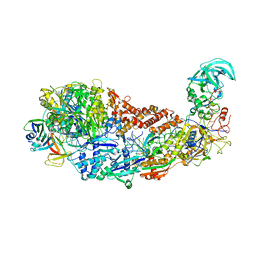 | | CRISPR-Cas type III-D effector complex | | Descriptor: | CRISPR RNA, Cas10, Cas7-2x, ... | | Authors: | Schwartz, E.A, Taylor, D.W. | | Deposit date: | 2023-03-30 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | RNA targeting and cleavage by the type III-Dv CRISPR effector complex.
Nat Commun, 15, 2024
|
|
8SJK
 
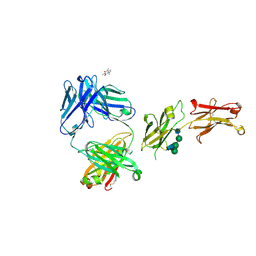 | | Pembrolizumab Caffeine crystal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY HEAVY CHAIN, ANTIBODY LIGHT CHAIN, ... | | Authors: | Larpent, P, Codan, L, Bothe, J.R, Stueber, D, Reichert, P, Fischmann, T, Su, Y, Pabit, S, Gupta, S, Iuzzolino, L, Cote, A. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Smaall angle X-ray scattering as powerful toolsfor phase and crystallinity assessment of monoclonal antibodies crystallites in support of batch crystallization and processing
To Be Published
|
|
8S9V
 
 | |
8S9X
 
 | |
2M1W
 
 | | TICAM-2 TIR domain | | Descriptor: | TIR domain-containing adapter molecule 2 | | Authors: | Enokizono, Y, Kumeta, H, Funami, K, Horiuchi, M, Sarmiento, J, Yamashita, K, Standley, D.M, Matsumoto, M, Seya, T, Inagaki, F. | | Deposit date: | 2012-12-07 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures and interface mapping of the TIR domain-containing adaptor molecules involved in interferon signaling.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HHR
 
 | | Crystal Structure of fatty acid alpha-dioxygenase (Arabidopsis thaliana) | | Descriptor: | Alpha-dioxygenase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Goulah, C.C, Zhu, G, Koszelak-Rosenblum, M, Malkowski, M.G. | | Deposit date: | 2012-10-10 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The crystal structure of alpha-Dioxygenase provides insight into diversity in the cyclooxygenase-peroxidase superfamily.
Biochemistry, 52, 2013
|
|
4HHS
 
 | | Crystal Structure of fatty acid alpha-dioxygenase (Arabidopsis thaliana) | | Descriptor: | Alpha-dioxygenase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Goulah, C.C, Zhu, G, Koszelak-Rosenblum, M, Malkowski, M.G. | | Deposit date: | 2012-10-10 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of alpha-Dioxygenase provides insight into diversity in the cyclooxygenase-peroxidase superfamily.
Biochemistry, 52, 2013
|
|
