1MI3
 
 | | 1.8 Angstrom structure of xylose reductase from Candida tenuis in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, xylose reductase | | Authors: | Kavanagh, K.L, Klimacek, M, Nidetzky, B, Wilson, D.K. | | Deposit date: | 2002-08-21 | | Release date: | 2003-08-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of xylose reductase bound to NAD+ and the basis for single and dual co-substrate specificity in family 2 aldo-keto reductases
Biochem.J., 373, 2003
|
|
1J9E
 
 | | Low Temperature (100K) Crystal Structure of Flavodoxin D. vulgaris S35C Mutant at 1.44 Angstrom Resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Artali, R, Bombieri, G, Meneghetti, F, Gilardi, G, Sadeghi, S.J, Cavazzini, D, Rossi, G.L. | | Deposit date: | 2001-05-25 | | Release date: | 2001-09-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Comparison of the refined crystal structures of wild-type (1.34 A) flavodoxin from Desulfovibrio vulgaris and the S35C mutant (1.44 A) at 100 K.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3E3K
 
 | | Structural characterization of a putative endogenous metal chelator in the periplasmic nickel transporter NikA (butane-1,2,4-tricarboxylate without nickel form) | | Descriptor: | (2R)-butane-1,2,4-tricarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Cherrier, M.V, Cavazza, C, Bochot, C, Lemaire, D, Fontecilla-Camps, J.C. | | Deposit date: | 2008-08-07 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of a putative endogenous metal chelator in the periplasmic nickel transporter NikA
Biochemistry, 47, 2008
|
|
2NW6
 
 | | Burkholderia cepacia lipase complexed with S-inhibitor | | Descriptor: | (1S)-1-(PHENOXYMETHYL)PROPYL METHYLPHOSPHONOCHLORIDOATE, CALCIUM ION, Lipase, ... | | Authors: | Luic, M, Stefanic, Z. | | Deposit date: | 2006-11-14 | | Release date: | 2007-12-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Combined X-ray diffraction and QM/MM study of the Burkholderia cepacia lipase-catalyzed secondary alcohol esterification
J.Phys.Chem.B, 112, 2008
|
|
2YBT
 
 | | Crystal structure of human acidic chitinase in complex with bisdionin C | | Descriptor: | 1,1'-PROPANE-1,3-DIYLBIS(3,7-DIMETHYL-3,7-DIHYDRO-1H-PURINE-2,6-DIONE), ACIDIC MAMMALIAN CHITINASE, GLYCEROL | | Authors: | Sutherland, T.E, Andersen, O.A, Betou, M, Eggleston, I.M, Maizels, R.M, van Aalten, D, Allen, J.E. | | Deposit date: | 2011-03-10 | | Release date: | 2011-06-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Analyzing airway inflammation with chemical biology: dissection of acidic mammalian chitinase function with a selective drug-like inhibitor.
Chem. Biol., 18, 2011
|
|
3S5O
 
 | |
3SN7
 
 | | Highly Potent, Selective, and Orally Active Phosphodiestarase 10A Inhibitors | | Descriptor: | 8-fluoro-6-methoxy-3,4-dimethyl-1-(3-methylpyridin-4-yl)imidazo[1,5-a]quinoxaline, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Parris, K.D. | | Deposit date: | 2011-06-28 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Highly Potent, Selective, and Orally Active Phosphodiesterase 10A Inhibitors.
J.Med.Chem., 54, 2011
|
|
4QTE
 
 | | Structure of ERK2 in complex with VTX-11e, 4-{2-[(2-CHLORO-4-FLUOROPHENYL)AMINO]-5-METHYLPYRIMIDIN-4-YL}-N-[(1S)-1-(3-CHLOROPHENYL)-2-HYDROXYETHYL]-1H-PYRROLE-2-CARBOXAMIDE | | Descriptor: | 1,2-ETHANEDIOL, 4-{2-[(2-chloro-4-fluorophenyl)amino]-5-methylpyrimidin-4-yl}-N-[(1S)-1-(3-chlorophenyl)-2-hydroxyethyl]-1H-pyrrole-2-carboxamide, CHLORIDE ION, ... | | Authors: | Chaikuad, A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A unique inhibitor binding site in ERK1/2 is associated with slow binding kinetics.
Nat.Chem.Biol., 10, 2014
|
|
3VU6
 
 | | Short peptide HIV entry inhibitor MT-SC22EK with a M-T hook | | Descriptor: | MTSC22, SULFATE ION, Transmembrane protein gp41 | | Authors: | Yao, X, Chong, H.H, Waltersperger, S, Wang, M.T, He, Y.X, Cui, S. | | Deposit date: | 2012-06-19 | | Release date: | 2012-12-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.324 Å) | | Cite: | Short-peptide fusion inhibitors with high potency against wild-type and enfuvirtide-resistant HIV-1
Faseb J., 27, 2013
|
|
2DMW
 
 | | Solution structure of the LONGIN domain of Synaptobrevin-like protein 1 | | Descriptor: | synaptobrevin-like 1 variant | | Authors: | Ohnishi, S, Sato, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-24 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the LONGIN domain of Synaptobrevin-like protein 1
To be Published
|
|
3EFT
 
 | | Crystal structure of the complex between Carbonic Anhydrase II and a spin-labeled sulfonamide incorporating TEMPO moiety | | Descriptor: | 3-chloro-4-{[(1-hydroxy-2,2,6,6-tetramethylpiperidin-4-yl)carbamothioyl]amino}benzenesulfonamide, Carbonic anhydrase 2, MERCURY (II) ION, ... | | Authors: | Temperini, C, Cecchi, A, Scozzafava, A, Supuran, C.T. | | Deposit date: | 2008-09-10 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dissecting the Inhibition Mechanism of Cytosolic versus Transmembrane Carbonic Anhydrases by ESR
J.Phys.Chem.B, 113, 2009
|
|
5RET
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102269 | | Descriptor: | 1-{4-[(3-chlorophenyl)methyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
1D1J
 
 | | CRYSTAL STRUCTURE OF HUMAN PROFILIN II | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, PROFILIN II, ... | | Authors: | Nodelman, I.M, Bowman, G.D, Lindberg, U, Schutt, C.E. | | Deposit date: | 1999-09-17 | | Release date: | 2000-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure determination of human profilin II: A comparative structural analysis of human profilins.
J.Mol.Biol., 294, 1999
|
|
1CZF
 
 | | ENDO-POLYGALACTURONASE II FROM ASPERGILLUS NIGER | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, POLYGALACTURONASE II, ZINC ION | | Authors: | van Santen, Y, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1999-09-02 | | Release date: | 1999-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | 1.68-A crystal structure of endopolygalacturonase II from Aspergillus niger and identification of active site residues by site-directed mutagenesis.
J.Biol.Chem., 274, 1999
|
|
3SGW
 
 | |
2LWB
 
 | | Structural model of BAD-1 repeat loop by NMR | | Descriptor: | Adhesin WI-1 | | Authors: | Brandhorst, T, Klein, B, Tonelli, M. | | Deposit date: | 2012-07-26 | | Release date: | 2013-07-31 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | Structure and function of a fungal adhesin that mimics thrombospondin-1 by binding heparin sulfate glycosaminoglycan and suppressing T cell activation via interaction with CD47
To be Published
|
|
3SL5
 
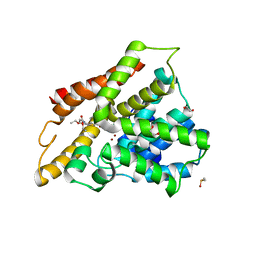 | | Crystal structure of the catalytic domain of PDE4D2 complexed with compound 10d | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2DL0
 
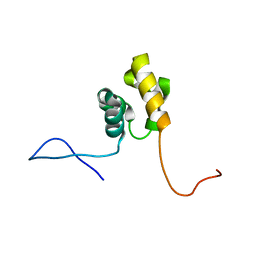 | | Solution structure of the SAM-domain of the SAM and SH3 domain containing protein 1 | | Descriptor: | SAM and SH3 domain-containing protein 1 | | Authors: | Goroncy, A.K, Sato, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-14 | | Release date: | 2006-10-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAM-domain of the SAM and SH3 domain containing protein 1
To be Published
|
|
3CFN
 
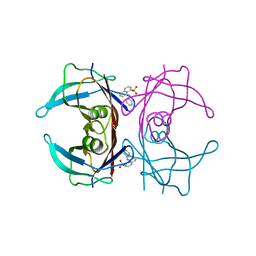 | |
1TG6
 
 | | Crystallography and mutagenesis point to an essential role for the N-terminus of human mitochondrial ClpP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, GLYCEROL, ... | | Authors: | Kang, S.G, Maurizi, M.R, Thompson, M, Mueser, T, Ahvazi, B. | | Deposit date: | 2004-05-28 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallography and mutagenesis point to an essential role for the N-terminus of human mitochondrial ClpP
J.Struct.Biol., 148, 2004
|
|
1K7H
 
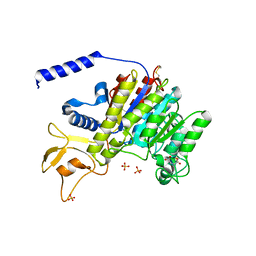 | | CRYSTAL STRUCTURE OF SHRIMP ALKALINE PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALKALINE PHOSPHATASE, MALEIC ACID, ... | | Authors: | De Backer, M.E, Mc Sweeney, S, Rasmussen, H.B, Riise, B.W, Lindley, P, Hough, E. | | Deposit date: | 2001-10-19 | | Release date: | 2002-07-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The 1.9 A Crystal Structure of Heat-Labile Shrimp Alkaline Phosphatase
J.Mol.Biol., 318, 2002
|
|
3NWO
 
 | |
4INS
 
 | | THE STRUCTURE OF 2ZN PIG INSULIN CRYSTALS AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | INSULIN (CHAIN A), INSULIN (CHAIN B), ZINC ION | | Authors: | Dodson, G.G, Dodson, E.J, Hodgkin, D.C, Isaacs, N.W, Vijayan, M. | | Deposit date: | 1989-07-10 | | Release date: | 1990-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of 2Zn pig insulin crystals at 1.5 A resolution.
Philos.Trans.R.Soc.London,Ser.B, 319, 1988
|
|
4A3X
 
 | | Structure of the N-terminal domain of the Epa1 adhesin (Epa1-Np) from the pathogenic yeast Candida glabrata, in complex with calcium and lactose | | Descriptor: | CALCIUM ION, EPA1P, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Ielasi, F.S, Decanniere, K, Willaert, R.G. | | Deposit date: | 2011-10-05 | | Release date: | 2012-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Epithelial Adhesin 1 (Epa1P) from the Human-Pathogenic Yeast Candida Glabrata : Structural and Functional Study of the Carbohydrate-Binding Domain
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1K4T
 
 | | HUMAN DNA TOPOISOMERASE I (70 KDA) IN COMPLEX WITH THE POISON TOPOTECAN AND COVALENT COMPLEX WITH A 22 BASE PAIR DNA DUPLEX | | Descriptor: | (S)-10-[(DIMETHYLAMINO)METHYL]-4-ETHYL-4,9-DIHYDROXY-1H-PYRANO[3',4':6,7]INOLIZINO[1,2-B]-QUINOLINE-3,14(4H,12H)-DIONE, 2-(1-DIMETHYLAMINOMETHYL-2-HYDROXY-8-HYDROXYMETHYL-9-OXO-9,11-DIHYDRO-INDOLIZINO[1,2-B]QUINOLIN-7-YL)-2-HYDROXY-BUTYRIC ACID, 5'-D(*(TGP)P*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', ... | | Authors: | Staker, B.L, Hjerrild, K, Feese, M.D, Behnke, C.A, Burgin Jr, A.B, Stewart, L.J. | | Deposit date: | 2001-10-08 | | Release date: | 2002-12-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The mechanism of topoisomerase I poisoning by a camptothecin analog
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
