5E84
 
 | | ATP-bound state of BiP | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Liu, Q, Yang, J, Nune, M, Zong, Y, Zhou, L. | | Deposit date: | 2015-10-13 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Close and Allosteric Opening of the Polypeptide-Binding Site in a Human Hsp70 Chaperone BiP.
Structure, 23, 2015
|
|
6T0E
 
 | | The glucuronoyl esterase OtCE15A S267A variant from Opitutus terrae in complex with benzyl D-glucuronoate and D-glucuronate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mazurkewich, S, Navarro Poulsen, J.C, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2019-10-03 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and biochemical studies of the glucuronoyl esteraseOtCE15A illuminate its interaction with lignocellulosic components.
J.Biol.Chem., 294, 2019
|
|
5EAL
 
 | | Crystal structure of human WDR5 in complex with compound 9h | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-~{N}-[2-(4-methylpiperazin-1-yl)-5-quinolin-3-yl-phenyl]benzamide, CHLORIDE ION, ... | | Authors: | DONG, A, DOMBROVSKI, L, SMIL, D, GETLIK, M, BOLSHAN, Y, WALKER, J.R, SENISTERRA, G, PODA, G, AL-AWAR, R, SCHAPIRA, M, VEDADI, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human WDR5 in complex with compound 9h
to be published
|
|
6SSI
 
 | | Structure of the pentameric ligand-gated ion channel ELIC in complex with a PAM nanobody | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, CALCIUM ION, ... | | Authors: | Ulens, C, Brams, M, Evans, G.L, Spurny, R, Govaerts, C, Pardon, E, Steyaert, J. | | Deposit date: | 2019-09-07 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Modulation of the Erwinia ligand-gated ion channel (ELIC) and the 5-HT 3 receptor via a common vestibule site.
Elife, 9, 2020
|
|
6H4F
 
 | | TarP-3RboP | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Guo, Y, Stehle, T. | | Deposit date: | 2018-07-21 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Methicillin-resistant Staphylococcus aureus alters cell wall glycosylation to evade immunity.
Nature, 563, 2018
|
|
6CEA
 
 | | Crystal structure of fragment 3-(quinolin-2-yl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(quinolin-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Ravichandran, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
6CEE
 
 | | Crystal structure of fragment 3-(1-Methyl-2-oxo-1,2-dihydroquinoxalin-3-yl)propionic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(4-methyl-3-oxo-3,4-dihydroquinoxalin-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Franzoni, I, Ravichandran, M, Lautens, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
6H7F
 
 | | Crystal structure of BauA, the Ferric preacinetobactin receptor from Acinetobacter baumannii in complex with Fe3+-Preacinetobactin-acinetobactin | | Descriptor: | (4~{S},5~{R})-2-[2,3-bis(oxidanyl)phenyl]-~{N}-[2-(1~{H}-imidazol-4-yl)ethyl]-5-methyl-~{N}-oxidanyl-4,5-dihydro-1,3-oxazole-4-carboxamide, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, ... | | Authors: | Moynie, L, Naismith, J.H. | | Deposit date: | 2018-07-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Preacinetobactin not acinetobactin is essential for iron uptake by the BauA transporter of the pathogenAcinetobacter baumannii.
Elife, 7, 2018
|
|
6SLN
 
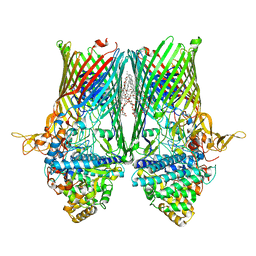 | | Structure of the RagAB peptide transporter | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Madej, M, Ranson, N.A, White, J.B.R. | | Deposit date: | 2019-08-20 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural and functional insights into oligopeptide acquisition by the RagAB transporter from Porphyromonas gingivalis.
Nat Microbiol, 5, 2020
|
|
6SNT
 
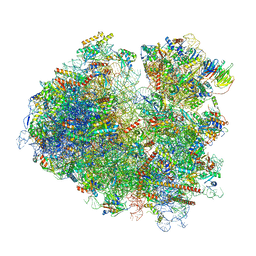 | | Yeast 80S ribosome stalled on SDD1 mRNA. | | Descriptor: | 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, 40S ribosomal protein S10-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Becker, T, Beckmann, R. | | Deposit date: | 2019-08-27 | | Release date: | 2020-03-04 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | RQT complex dissociates ribosomes collided on endogenous RQC substrate SDD1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6H77
 
 | | E1 enzyme for ubiquitin like protein activation in complex with UBL | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Soudah, N, Padala, P, Hassouna, F, Mashahreh, B, Lebedev, A.A, Isupov, M.N, Cohen-Kfir, E, Wiener, R. | | Deposit date: | 2018-07-30 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An N-Terminal Extension to UBA5 Adenylation Domain Boosts UFM1 Activation: Isoform-Specific Differences in Ubiquitin-like Protein Activation.
J.Mol.Biol., 431, 2019
|
|
6SW9
 
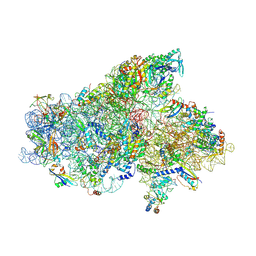 | | IC2A model of cryo-EM structure of a full archaeal ribosomal translation initiation complex devoid of aIF1 in P. abyssi | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Coureux, P.-D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of translation initiation.
Commun Biol, 3, 2020
|
|
4TNW
 
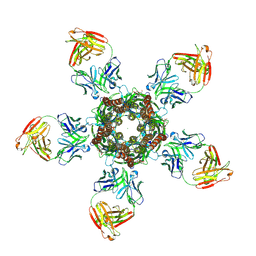 | | C. elegans glutamate-gated chloride channel (GluCl) in complex with Fab and POPC in a lipid-modulated conformation | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Avermectin-sensitive glutamate-gated chloride channel GluCl alpha, ... | | Authors: | Althoff, T, Hibbs, R.E, Banerjee, S, Gouaux, E. | | Deposit date: | 2014-06-05 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors.
Nature, 512, 2014
|
|
6HCQ
 
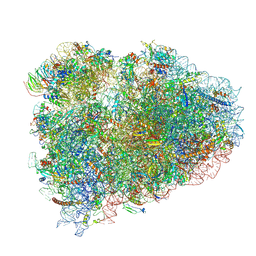 | | Structure of the rabbit collided di-ribosome (collided monosome) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-16 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
4S0R
 
 | | Structure of GS-TnrA complex | | Descriptor: | GLUTAMINE, Glutamine synthetase, MAGNESIUM ION, ... | | Authors: | Schumacher, M.A, Chinnam, N.G, Cuthbert, B, Tonthat, N.K. | | Deposit date: | 2015-01-04 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of regulatory machinery reveal novel molecular mechanisms controlling B. subtilis nitrogen homeostasis.
Genes Dev., 29, 2015
|
|
1MCZ
 
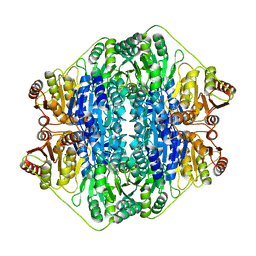 | | BENZOYLFORMATE DECARBOXYLASE FROM PSEUDOMONAS PUTIDA COMPLEXED WITH AN INHIBITOR, R-MANDELATE | | Descriptor: | (R)-MANDELIC ACID, BENZOYLFORMATE DECARBOXYLASE, MAGNESIUM ION, ... | | Authors: | Polovnikova, E.S, Bera, A.K, Hasson, M.S. | | Deposit date: | 2002-08-06 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Kinetic Analysis of Catalysis by a Thiamin
Diphosphate-Dependent Enzyme, Benzoylformate Decarboxylase
Biochemistry, 42, 2003
|
|
6SWA
 
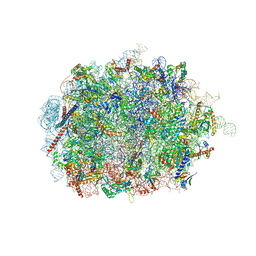 | | Mus musculus brain neocortex ribosome 60S bound to Ebp1 | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kraushar, M.L, Sprink, T. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-30 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Protein Synthesis in the Developing Neocortex at Near-Atomic Resolution Reveals Ebp1-Mediated Neuronal Proteostasis at the 60S Tunnel Exit.
Mol.Cell, 81, 2021
|
|
6HUV
 
 | | Yeast 20S proteasome with human beta2c (S171G) in complex with 39 | | Descriptor: | (2~{S})-~{N}-[(3~{S},4~{R})-1-cyclohexyl-5-methyl-4,5-bis(oxidanyl)hexan-3-yl]-3-(4-methoxyphenyl)-2-[[(2~{S})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-09 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
6HJD
 
 | | Cholera toxin classical B-pentamer in complex with Lewis-x | | Descriptor: | BICINE, CALCIUM ION, Cholera toxin B subunit, ... | | Authors: | Krengel, U, Heim, J.B. | | Deposit date: | 2018-09-03 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structures of cholera toxin in complex with fucosylated receptors point to importance of secondary binding site.
Sci Rep, 9, 2019
|
|
6HW5
 
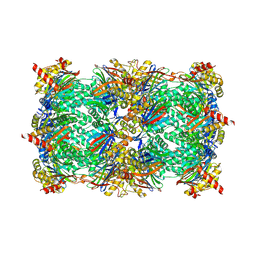 | | Yeast 20S proteasome in complex with 18 | | Descriptor: | (2~{S})-~{N}-[2-[[(2~{S})-1-[4-(aminomethyl)phenyl]-4-methylsulfonyl-butan-2-yl]amino]-2-oxidanylidene-ethyl]-2-[[(2~{S})-2-azido-3-phenyl-propanoyl]amino]-4-methyl-pentanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
4TT0
 
 | |
6HK6
 
 | | Human RIOK2 bound to inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-naphthalen-2-yl-~{N}-pyridin-2-yl-ethanamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Wang, J, Krojer, T, Bountra, C, Edwards, A.M, Arrowsmith, C, Knapp, S, Elkins, J.M. | | Deposit date: | 2018-09-05 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of human RIOK2 bound to a specific inhibitor.
Open Biology, 9, 2019
|
|
5E1D
 
 | | NTMT1 in complex with YPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
5E1O
 
 | | Crystal structure of NTMT1 in complex with RPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
6HXH
 
 | | Structure of the human ATP citrate lyase holoenzyme in complex with citrate, coenzyme A and Mg.ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase,Human ATP citrate lyase, CITRATE ANION, ... | | Authors: | Verstraete, K, Verschueren, K. | | Deposit date: | 2018-10-17 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of ATP citrate lyase and the origin of citrate synthase in the Krebs cycle.
Nature, 568, 2019
|
|
