2QNC
 
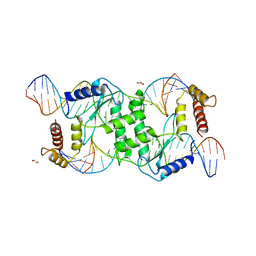 | | Crystal structure of T4 Endonuclease VII N62D mutant in complex with a DNA Holliday junction | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*DAP*DGP*DGP*DCP*DCP*DTP*DAP*DGP*DCP*DGP*DTP*DCP*DCP*DGP*DGP*DAP*DAP*DTP*DTP*DCP*DTP*DTP*DCP*DG)-3'), DNA (5'-D(*DCP*DAP*DCP*DAP*DTP*DCP*DGP*DAP*DTP*DGP*DGP*DAP*DGP*DCP*DCP*DGP*DCP*DTP*DAP*DGP*DGP*DCP*DCP*DT)-3'), ... | | Authors: | Biertumpfel, C, Yang, W, Suck, D. | | Deposit date: | 2007-07-18 | | Release date: | 2008-01-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of T4 endonuclease VII resolving a Holliday junction.
Nature, 449, 2007
|
|
3B9G
 
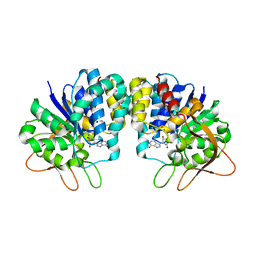 | | Crystal structure of loop deletion mutant of Trypanosoma vivax nucleoside hydrolase (3GTvNH) in complex with ImmH | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, IAG-nucleoside hydrolase, ... | | Authors: | Vandemeulebroucke, A, De Vos, S, Van Holsbeke, E, Steyaert, J, Versees, W. | | Deposit date: | 2007-11-05 | | Release date: | 2008-04-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Flexible Loop as a Functional Element in the Catalytic Mechanism of Nucleoside Hydrolase from Trypanosoma vivax.
J.Biol.Chem., 283, 2008
|
|
4AS2
 
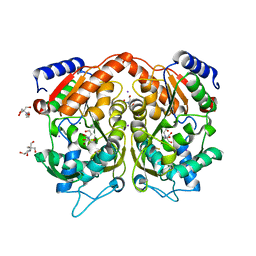 | | Pseudomonas Aeruginosa Phosphorylcholine Phosphatase. Monoclinic form | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, IODIDE ION, ... | | Authors: | Infantes, L, Otero, L.H, Albert, A. | | Deposit date: | 2012-04-27 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The Structural Domains of Pseudomonas Aeruginosa Phosphorylcholine Phosphatase Cooperate in Substrate Hydrolysis: 3D Structure and Enzymatic Mechanism.
J.Mol.Biol., 423, 2012
|
|
1W91
 
 | |
1W9I
 
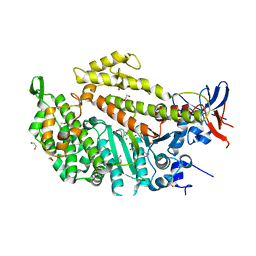 | | Myosin II Dictyostelium discoideum motor domain S456Y bound with MgADP-BeFx | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Morris, C.A, Coureux, P.-D, Wells, A.L, Houdusse, A, Sweeney, H.L. | | Deposit date: | 2004-10-13 | | Release date: | 2006-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Function Analysis of Myosin II Backdoor Mutants
To be Published
|
|
6LSN
 
 | | Crystal structure of tubulin-inhibitor complex | | Descriptor: | 2-(1-methylindol-5-yl)-7-(3,4,5-trimethoxyphenyl)pyrazolo[1,5-a]pyrimidine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Gang, L, Wang, Y.X, Cheng, J.J. | | Deposit date: | 2020-01-17 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Design, Synthesis, and Bioevaluation of Pyrazolo[1,5-a]Pyrimidine Derivatives as Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site with Potent Anticancer Activities
To Be Published
|
|
1Q35
 
 | | Crystal Structure of Pasteurella haemolytica Apo Ferric ion-Binding Protein A | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, iron binding protein FbpA | | Authors: | Shouldice, S.R, Dougan, D.R, Skene, R.J, Snell, G, Scheibe, D, Williams, P.A, Kirby, S, McRee, D.E, Schryvers, A.B, Tari, L.W. | | Deposit date: | 2003-07-28 | | Release date: | 2003-11-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of Pasteurella haemolytica ferric ion-binding protein A reveals a novel class of bacterial iron-binding proteins
J.Biol.Chem., 278, 2003
|
|
3BJK
 
 | | Crystal structure of HI0827, a hexameric broad specificity acyl-coenzyme A thioesterase: The Asp44Ala mutant enzyme | | Descriptor: | 1,2-ETHANEDIOL, Acyl-CoA thioester hydrolase HI0827, CITRIC ACID | | Authors: | Willis, M.A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2007-12-04 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of YciA from Haemophilus influenzae (HI0827), a Hexameric Broad Specificity Acyl-Coenzyme A Thioesterase.
Biochemistry, 47, 2008
|
|
4Q9O
 
 | | Crystal structure of Upps + inhibitor | | Descriptor: | 3-(2-chlorophenyl)-5-methyl-N-[4-(propan-2-yl)phenyl]-1,2-oxazole-4-carboxamide, Isoprenyl transferase, SULFATE ION | | Authors: | Liu, S, Qiu, X. | | Deposit date: | 2014-05-01 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and structural characterization of an allosteric inhibitor of bacterial cis-prenyltransferase.
Protein Sci., 24, 2015
|
|
5CYQ
 
 | | HIV-1 reverse transcriptase complexed with 4-bromopyrazole | | Descriptor: | 4-bromo-1H-pyrazole, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, BROMIDE ION, ... | | Authors: | Bauman, J.D, Arnold, E. | | Deposit date: | 2015-07-30 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Rapid experimental SAD phasing and hot-spot identification with halogenated fragments.
IUCrJ, 3, 2016
|
|
3M3M
 
 | | Crystal structure of glutathione S-transferase from Pseudomonas fluorescens [Pf-5] | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase, ... | | Authors: | Bagaria, A, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-16 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of glutathione S-transferase from Pseudomonas fluorescens [Pf-5]
To be Published
|
|
7AY9
 
 | | Crystal structure of CK2 bound by compound 7 | | Descriptor: | 1,2-ETHANEDIOL, 7-(cyclopropylamino)-5-(5-(6-oxo-1,6-dihydropyridin-3-yl)-1-(2-(piperidin-1-yl)ethyl)-1H-1,2,3-triazol-4-yl)pyrazolo[1,5-a]pyrimidine-3-carbonitrile, Casein kinase II subunit alpha, ... | | Authors: | Ferguson, A, Collie, G.W. | | Deposit date: | 2020-11-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Metadynamics simulations of CK2 compound unbinding to understand slow dissociation kinetics.
To Be Published
|
|
7AOU
 
 | | The Fk1 domain of FKBP51 in complex with (2'R,5'S,12'R)-12'-cyclohexyl-2'-[2-(3,4-dimethoxyphenyl)ethyl]-3',19'-dioxa-10',13',16'-triazaspiro[cyclopropane-1,15'- tricyclo[18.3.1.0-5,10]tetracosane]-1'(24'),20',22'-triene-4',11',14',17'-tetrone | | Descriptor: | (2'R,5'S,12'R)-12'-cyclohexyl-2'-[2-(3,4-dimethoxyphenyl)ethyl]-3',19'-dioxa-10',13',16'-triazaspiro[cyclopropane-1,15'- tricyclo[18.3.1.0-5,10]tetracosane]-1'(24'),20',22'-triene-4',11',14',17'-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Voll, M.A, Meyners, C, Heymann, T, Merz, S, Purder, P, Bracher, A, Hausch, F. | | Deposit date: | 2020-10-15 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Macrocyclic FKBP51 Ligands Define a Transient Binding Mode with Enhanced Selectivity.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5WUM
 
 | |
6D28
 
 | | Crystal Structure of the N-domain of the ER Hsp90 chaperone GRP94 in complex with the specific ligand NECA | | Descriptor: | 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, Endoplasmin, N-ETHYL-5'-CARBOXAMIDO ADENOSINE | | Authors: | Soldano, K.L, Jivan, A, Nicchitta, C.V, Gewirth, D.T. | | Deposit date: | 2018-04-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the N-terminal domain of GRP94. Basis for ligand specificity and regulation.
J. Biol. Chem., 278, 2003
|
|
5DFF
 
 | | Human APE1 product complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-08-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Capturing snapshots of APE1 processing DNA damage.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5WFL
 
 | | Kelch domain of human Keap1 in open unliganded conformation | | Descriptor: | Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Carolan, J.P, Lynch, A.J, Allen, K.N. | | Deposit date: | 2017-07-12 | | Release date: | 2018-09-19 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Interaction Energetics and Druggability of the Protein-Protein Interaction between Kelch-like ECH-Associated Protein 1 (KEAP1) and Nuclear Factor Erythroid 2 Like 2 (Nrf2).
Biochemistry, 59, 2020
|
|
5WLT
 
 | | Carbonic Anhydrase IX-mimic in complex with aryloxy-2-hydroxypropylammine sulfonamide | | Descriptor: | 4-{(2S)-2-hydroxy-3-[(propan-2-yl)amino]propoxy}benzene-1-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Lomelino, C.L, Andring, J.T, McKenna, R. | | Deposit date: | 2017-07-27 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Discovery of beta-Adrenergic Receptors Blocker-Carbonic Anhydrase Inhibitor Hybrids for Multitargeted Antiglaucoma Therapy.
J. Med. Chem., 61, 2018
|
|
7KN1
 
 | |
7E2E
 
 | | Crystal structure of the Estrogen-Related Receptor alpha (ERRalpha) ligand-binding domain (LBD) in complex with an agonist DS45500853 and a PGC-1alpha peptide | | Descriptor: | 1-[4-(3-tert-butyl-4-oxidanyl-phenoxy)phenyl]ethanone, IODIDE ION, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, ... | | Authors: | Ito, S, Shinozuka, T, Kimura, T, Izumi, M, Wakabayashi, K. | | Deposit date: | 2021-02-05 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of a Novel Class of ERR alpha Agonists.
Acs Med.Chem.Lett., 12, 2021
|
|
7VNE
 
 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with a human single domain antibody n3113.1 (UUU-state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
3FMD
 
 | | Crystal Structure of Human Haspin with an Isoquinoline ligand | | Descriptor: | 1,2-ETHANEDIOL, N-[2-(4-BROMOCINNAMYLAMINO)ETHYL]-5-ISOQUINOLINE SULFONAMIDE, NICKEL (II) ION, ... | | Authors: | Filippakopoulos, P, Eswaran, J, Keates, T, Burgess-Brown, N, Fedorov, O, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-21 | | Release date: | 2008-12-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Haspin with an Isoquinoline ligand
To be Published
|
|
7K6M
 
 | | Crystal structure of PI3Kalpha selective Inhibitor PF-06843195 | | Descriptor: | 2,2-difluoroethyl (3S)-3-{[2'-amino-5-fluoro-2-(morpholin-4-yl)[4,5'-bipyrimidin]-6-yl]amino}-3-(hydroxymethyl)pyrrolidine-1-carboxylate, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Chen, P, Brooun, A, Deng, Y.L, Grodsky, N, Kaiser, S.E. | | Deposit date: | 2020-09-21 | | Release date: | 2021-01-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structure-Based Drug Design and Synthesis of PI3K alpha-Selective Inhibitor (PF-06843195).
J.Med.Chem., 64, 2021
|
|
6MBB
 
 | | Human Bfl-1 in complex with the designed peptide dF1 | | Descriptor: | Bcl-2-related protein A1, dF1 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
7VNC
 
 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with a human single domain antibody n3113 (UDD-state, state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
