6MW3
 
 | | EM structure of Bacillus subtilis ribonucleotide reductase inhibited filament composed of NrdE alpha subunit and NrdF beta subunit with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Ribonucleoside-diphosphate reductase, Ribonucleoside-diphosphate reductase NrdF beta subunit | | Authors: | Thomas, W.C, Bacik, J.P, Kaelber, J.T, Ando, N. | | Deposit date: | 2018-10-29 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.65 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
6N84
 
 | | MBP-fusion protein of transducin-alpha residues 327-350 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Guanine nucleotide-binding protein G(t) subunit alpha-2, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Srivastava, D, Gakhar, L, Artemyev, N.O. | | Deposit date: | 2018-11-28 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural underpinnings of Ric8A function as a G-protein alpha-subunit chaperone and guanine-nucleotide exchange factor.
Nat Commun, 10, 2019
|
|
5T05
 
 | | Crystal structure of heparan sulfate 6-O-sulfotransferase with bound PAP and IdoA2S containing hexasaccharide substrate | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Pedersen, L.C, Moon, A.F, krahn, J.M, Liu, J. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structure Based Substrate Specificity Analysis of Heparan Sulfate 6-O-Sulfotransferases.
ACS Chem. Biol., 12, 2017
|
|
5T6R
 
 | | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis: 60S-Nmd3 Complex | | Descriptor: | 25S Ribosomal RNA, 5.8S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Malyutin, A.G, Musalgaonkar, S, Patchett, S, Frank, J, Johnson, A.W. | | Deposit date: | 2016-09-01 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis.
EMBO J., 36, 2017
|
|
3F5F
 
 | | Crystal structure of heparan sulfate 2-O-sulfotransferase from gallus gallus as a maltose binding protein fusion. | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Maltose-binding periplasmic protein, Heparan sulfate 2-O-sulfotransferase 1, ... | | Authors: | Bethea, H.N, Xu, D, Liu, J, Pedersen, L.C. | | Deposit date: | 2008-11-03 | | Release date: | 2008-12-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Redirecting the substrate specificity of heparan sulfate 2-O-sulfotransferase by structurally guided mutagenesis.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6M4V
 
 | | Crystal structure of MBP fused split FKBP in complex with rapamycin | | Descriptor: | GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP1A, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, ... | | Authors: | Kikuchi, M, Wu, D, Inoue, T, Umehara, T. | | Deposit date: | 2020-03-09 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Rational design and implementation of a chemically inducible heterotrimerization system.
Nat.Methods, 17, 2020
|
|
6O6D
 
 | |
5T03
 
 | | Crystal structure of heparan sulfate 6-O-sulfotransferase with bound PAP and glucuronic acid containing hexasaccharide substrate | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Pedersen, L.C, Moon, A.F, Krahn, J.M, Liu, J. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure Based Substrate Specificity Analysis of Heparan Sulfate 6-O-Sulfotransferases.
ACS Chem. Biol., 12, 2017
|
|
1SVX
 
 | | Crystal structure of a designed selected Ankyrin Repeat protein in complex with the Maltose Binding Protein | | Descriptor: | Ankyrin Repeat Protein off7, Maltose-binding periplasmic protein | | Authors: | Binz, H.K, Amstutz, P, Kohl, A, Stumpp, M.T, Briand, C, Forrer, P, Gruetter, M.G, Plueckthun, A. | | Deposit date: | 2004-03-30 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | High-affinity binders selected from designed ankyrin repeat protein libraries
NAT.BIOTECHNOL., 22, 2004
|
|
5TJ4
 
 | |
3G7W
 
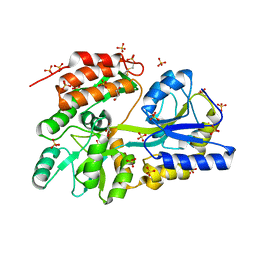 | | Islet Amyloid Polypeptide (IAPP or Amylin) Residues 1 to 22 fused to Maltose Binding Protein | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein, Islet amyloid polypeptide fusion protein, ... | | Authors: | Wiltzius, J.J.W, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2009-02-11 | | Release date: | 2009-06-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Atomic structures of IAPP (amylin) fusions suggest a mechanism for fibrillation and the role of insulin in the process
Protein Sci., 18, 2009
|
|
2KLF
 
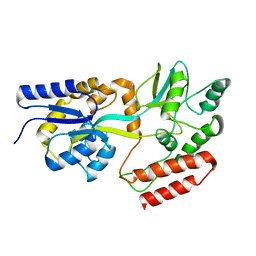 | | PERE NMR structure of maltodextrin-binding protein | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Madl, T, Bermel, W, Zangger, K. | | Deposit date: | 2009-07-02 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Use of Relaxation Enhancements in a Paramagnetic Environment for the Structure Determination of Proteins Using NMR Spectroscopy
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
5TTD
 
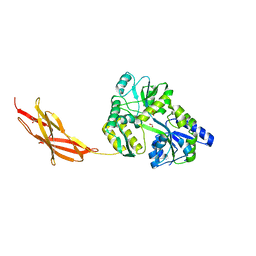 | | Minor pilin FctB from S. pyogenes with engineered intramolecular isopeptide bond | | Descriptor: | FORMIC ACID, Maltose-binding periplasmic protein,Pilin isopeptide linkage domain protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Young, P.G, Kwon, H, Squire, C.J, Baker, E.N. | | Deposit date: | 2016-11-02 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering a Lys-Asn isopeptide bond into an immunoglobulin-like protein domain enhances its stability.
Sci Rep, 7, 2017
|
|
2MV0
 
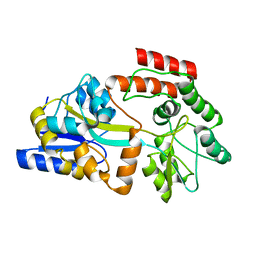 | | Solution NMR Structure of Maltose-binding protein from Escherichia coli, Northeast Structural Genomics Consortium (NESG) Target ER690 | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Rossi, P, Lange, O.F, Sgourakis, N.G, Song, Y, Lee, H, Aramini, J.M, Ertekin, A, Xiao, R, Acton, T.B, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5TIB
 
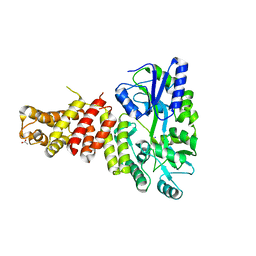 | | Gasdermin-B C-terminal domain containing the polymorphism residues Arg299:Ser306 fused to maltose binding protein | | Descriptor: | ACETATE ION, SODIUM ION, Sugar ABC transporter substrate-binding protein,Gasdermin-B, ... | | Authors: | Chao, K, Herzberg, O. | | Deposit date: | 2016-10-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human Gasdermin-B and disease: Sulfatide Binding, Caspase cleavage, and Structural impact of Asthma- and IBS-Associated Polymorphism
Proc.Natl.Acad.Sci.Usa, 2017
|
|
2N44
 
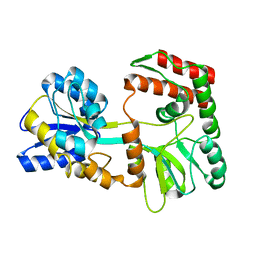 | | EC-NMR Structure of Escherichia coli Maltose-binding protein Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target ER690 | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-16 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
1T0K
 
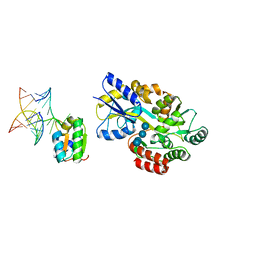 | | Joint X-ray and NMR Refinement of Yeast L30e-mRNA complex | | Descriptor: | 5'-R(*G*GP*AP*CP*GP*CP*AP*GP*AP*GP*AP*UP*GP*GP*UP*C)-3', 5'-R(*GP*AP*CP*CP*GP*GP*AP*GP*UP*GP*UP*CP*C)-3', 60S ribosomal protein L30, ... | | Authors: | Chao, J.A, Williamson, J.R. | | Deposit date: | 2004-04-09 | | Release date: | 2004-07-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Joint X-Ray and NMR Refinement of the Yeast L30e-mRNA Complex
Structure, 12, 2004
|
|
1PEB
 
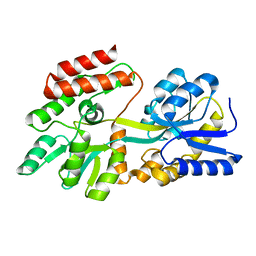 | | LIGAND-FREE HIGH-AFFINITY MALTOSE-BINDING PROTEIN | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Telmer, P.G, Shilton, B.H. | | Deposit date: | 2003-05-21 | | Release date: | 2003-08-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into the Conformational Equilibria of Maltose-binding Protein by Analysis of High Affinity Mutants.
J.Biol.Chem., 278, 2003
|
|
2N45
 
 | | EC-NMR Structure of Escherichia coli Maltose-binding protein Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data with a second set of RDC data simulated for an alternative alignment tensor. Northeast Structural Genomics Consortium target ER690 | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
3WOA
 
 | |
7O2W
 
 | | Structure of the C9orf72-SMCR8 complex | | Descriptor: | Guanine nucleotide exchange protein SMCR8,Guanine nucleotide exchange protein SMCR8,Maltose/maltodextrin-binding periplasmic protein, Ubiquitin-like protein SMT3,Guanine nucleotide exchange C9orf72 | | Authors: | Noerpel, J, Cavadini, S, Schenk, A.D, Graff-Meyer, A, Chao, J, Bhaskar, V. | | Deposit date: | 2021-03-31 | | Release date: | 2021-07-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the human C9orf72-SMCR8 complex reveals a multivalent protein interaction architecture.
Plos Biol., 19, 2021
|
|
7NZM
 
 | | Cryo-EM structure of pre-dephosphorylation complex of phosphorylated eIF2alpha with trapped holophosphatase (PP1A_D64A/PPP1R15A/G-actin/DNase I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yan, Y, Hardwick, S, Ron, D. | | Deposit date: | 2021-03-24 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Higher-order phosphatase-substrate contacts terminate the integrated stress response.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8W23
 
 | | Cryo-EM structure of human tankyrase 2 SAM-PARP filament bound to compound, TDI-2804 (consensus map). | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Poly [ADP-ribose] polymerase tankyrase-2, N-{2-[4-(2-hydroxypropan-2-yl)phenyl]-4-oxo-1,4-dihydroquinazolin-7-yl}-4-methoxy-6-phenylpyridine-3-carboxamide, ZINC ION | | Authors: | Malone, B.F, Zimmerman, J.L, Dow, L.E, Hite, R.K. | | Deposit date: | 2024-02-19 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | A potent and selective TNKS2 inhibitor for tumor-selective WNT suppression.
Biorxiv, 2025
|
|
8W25
 
 | | Cryo-EM structure of human tankyrase 2 SAM-PARP filament bound to compound, TDI-2804 (focused refinement map). | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Poly [ADP-ribose] polymerase tankyrase-2, N-{2-[4-(2-hydroxypropan-2-yl)phenyl]-4-oxo-1,4-dihydroquinazolin-7-yl}-4-methoxy-6-phenylpyridine-3-carboxamide, ZINC ION | | Authors: | Malone, B.F, Zimmerman, J.L, Dow, L.E, Hite, R.K. | | Deposit date: | 2024-02-20 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | A potent and selective TNKS2 inhibitor for tumor-selective WNT suppression.
Biorxiv, 2025
|
|
7NVM
 
 | | Human TRiC complex in closed state with nanobody Nb18, actin and PhLP2A bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Actin, ... | | Authors: | Kelly, J.J, Chi, G, Bulawa, C, Paavilainen, V.O, Bountra, C, Huiskonen, J.T, Yue, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Snapshots of actin and tubulin folding inside the TRiC chaperonin.
Nat.Struct.Mol.Biol., 29, 2022
|
|
