6UTE
 
 | | Crystal structure of Z032 Fab in complex with WNV EDIII | | Descriptor: | Envelope domain III, GLYCEROL, Z032 Fab heavy chain, ... | | Authors: | Esswein, S.R, Gristick, H.B, Keeffe, J.R, Bjorkman, P.J. | | Deposit date: | 2019-10-29 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for Zika envelope domain III recognition by a germline version of a recurrent neutralizing antibody.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UOE
 
 | |
6UMX
 
 | | Structural basis for specific inhibition of extracellular activation of pro/latent myostatin by SRK-015 | | Descriptor: | GL29H4-16 Fab Heavy Chain,GL29H4-16 Fab Heavy Chain, GL29H4-16 Fab Light Chain,GL29H4-16 Fab Light Chain, GLYCEROL, ... | | Authors: | Dagbay, K.B, Treece, E, Streich Jr, F.C, Jackson, J.W, Faucette, R.R, Nikiforov, A, Lin, S.C, Bostion, C.J, Nicholls, S.B, Capili, A.D, Carven, G.J. | | Deposit date: | 2019-10-10 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis of specific inhibition of extracellular activation of pro- or latent myostatin by the monoclonal antibody SRK-015.
J.Biol.Chem., 295, 2020
|
|
6UMV
 
 | | Human apo PD-1 double mutant | | Descriptor: | CHLORIDE ION, Programmed cell death protein 1 | | Authors: | Tang, S, Kim, P.S. | | Deposit date: | 2019-10-10 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.424 Å) | | Cite: | A high-affinity human PD-1/PD-L2 complex informs avenues for small-molecule immune checkpoint drug discovery.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6UMT
 
 | | High-affinity human PD-1 PD-L2 complex | | Descriptor: | MAGNESIUM ION, Programmed cell death 1 ligand 2, Programmed cell death protein 1 | | Authors: | Tang, S, Kim, P.S. | | Deposit date: | 2019-10-10 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | A high-affinity human PD-1/PD-L2 complex informs avenues for small-molecule immune checkpoint drug discovery.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6UMU
 
 | | Human apo PD-1 triple mutant | | Descriptor: | CHLORIDE ION, Programmed cell death protein 1 | | Authors: | Tang, S, Kim, P.S. | | Deposit date: | 2019-10-10 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.183 Å) | | Cite: | A high-affinity human PD-1/PD-L2 complex informs avenues for small-molecule immune checkpoint drug discovery.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6UGU
 
 | | Crystal structure of the Fab fragment of anti-TNFa antibody infliximab (Remicade) in a C-centered orthorhombic crystal form, Lot C | | Descriptor: | PF06438179 Fab Heavy Chain, PF06438179 Fab Light Chain | | Authors: | Lerch, T.F, Sharpe, P, Mayclin, S.J, Edwards, T.E, Polleck, S, Rouse, J.C, Conlan, H.D. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of PF-06438179/GP1111, an Infliximab Biosimilar.
BioDrugs, 34, 2020
|
|
6UGS
 
 | | Crystal structure of the Fab fragment of PF06438179/GP1111 an infliximab biosimilar in a C-centered orthorhombic crystal form, Lot A | | Descriptor: | Infliximab (Remicade) Fab Heavy Chain, Infliximab (Remicade) Fab Light Chain | | Authors: | Lerch, T.F, Sharpe, P, Mayclin, S.J, Edwards, T.E, Polleck, S, Rouse, J.C, Conlan, H. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of PF-06438179/GP1111, an Infliximab Biosimilar.
BioDrugs, 34, 2020
|
|
6UGT
 
 | | Crystal structure of the Fab fragment of PF06438179/GP1111 an infliximab biosimilar in a I-centered orthorhombic crystal form, Lot A | | Descriptor: | 1,2-ETHANEDIOL, PF06438179 Fab Heavy Chain, PF06438179 Fab Light Chain | | Authors: | Lerch, T.F, Sharpe, P, Mayclin, S.J, Edwards, T.E, Polleck, S, Rouse, J.C, Conlan, H.D. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of PF-06438179/GP1111, an Infliximab Biosimilar.
BioDrugs, 34, 2020
|
|
6UGV
 
 | | Crystal structure of the Fab fragment of anti-TNFa antibody infliximab (Remicade) in a I-centered orthorhombic crystal form, Lot C | | Descriptor: | 1,2-ETHANEDIOL, Infliximab Fab Heavy Chain, Infliximab Fab Light Chain, ... | | Authors: | Lerch, T.F, Sharpe, P, Mayclin, S.J, Edwards, T.E, Polleck, S, Rouse, J.C, Conlan, H.D. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of PF-06438179/GP1111, an Infliximab Biosimilar.
BioDrugs, 34, 2020
|
|
6UE7
 
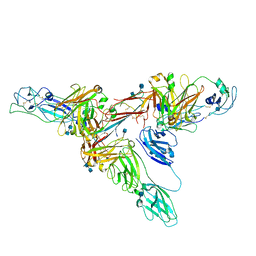 | | Structure of dimeric sIgA complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Kumar, N, Arthur, C.P, Ciferri, C, Matsumoto, M.L. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the secretory immunoglobulin A core.
Science, 367, 2020
|
|
6UEA
 
 | | Structure of pentameric sIgA complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Kumar, N, Arthur, C.P, Ciferri, C, Matsumoto, M.L. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the secretory immunoglobulin A core.
Science, 367, 2020
|
|
6UE9
 
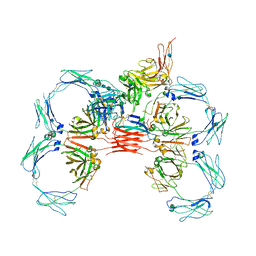 | | Structure of tetrameric sIgA complex (Class 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Kumar, N, Arthur, C.P, Ciferri, C, Matsumoto, M.L. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the secretory immunoglobulin A core.
Science, 367, 2020
|
|
6UE8
 
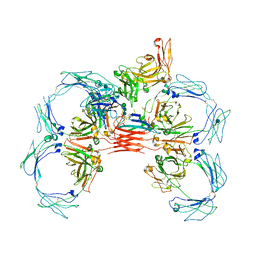 | | Structure of tetrameric sIgA complex (Class 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Kumar, N, Arthur, C.P, Ciferri, C, Matsumoto, M.L. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the secretory immunoglobulin A core.
Science, 367, 2020
|
|
6UDK
 
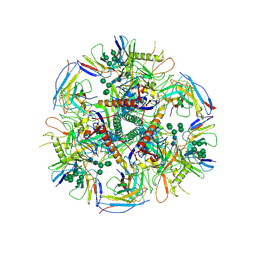 | | HIV-1 bNAb 1-55 in complex with modified BG505 SOSIP-based immunogen RC1 and 10-1074 | | Descriptor: | 1-55 Fab Heavy Chain, 1-55 Fab Light Chain, 10-1074 Fab Heavy Chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2019-09-19 | | Release date: | 2020-01-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Restriction of HIV-1 Escape by a Highly Broad and Potent Neutralizing Antibody.
Cell, 180, 2020
|
|
6UDJ
 
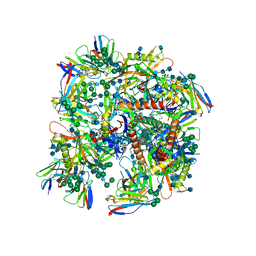 | | HIV-1 bNAb 1-18 in complex with BG505 SOSIP.664 and 10-1074 | | Descriptor: | 1-18 Fab Heavy Chain, 1-18 Fab Light Chain, 10-1074 Fab Heavy Chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2019-09-19 | | Release date: | 2020-01-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Restriction of HIV-1 Escape by a Highly Broad and Potent Neutralizing Antibody.
Cell, 180, 2020
|
|
6UCE
 
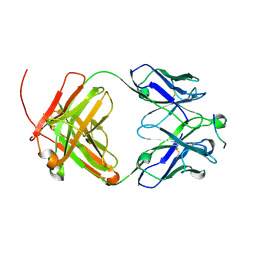 | |
6UCF
 
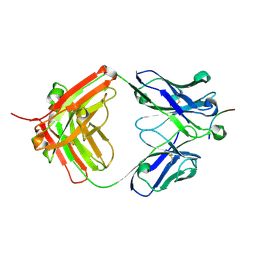 | |
6UC5
 
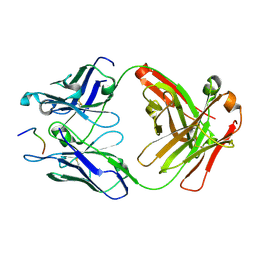 | | Fab397 in complex with NPNA peptide | | Descriptor: | Fab397 heavy chain, Fab397 light chain, NPNA peptide | | Authors: | Pholcharee, T, Oyen, D, Wilson, I.A. | | Deposit date: | 2019-09-13 | | Release date: | 2020-01-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Diverse Antibody Responses to Conserved Structural Motifs in Plasmodium falciparum Circumsporozoite Protein.
J.Mol.Biol., 432, 2020
|
|
6KXS
 
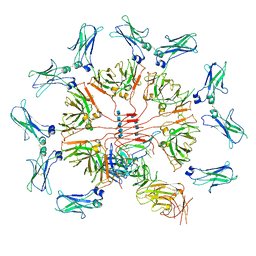 | | Cryo-EM structure of human IgM-Fc in complex with the J chain and the ectodomain of pIgR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Li, Y, Wang, G, Xiao, J. | | Deposit date: | 2019-09-12 | | Release date: | 2020-02-05 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into immunoglobulin M.
Science, 367, 2020
|
|
6UBI
 
 | |
6SRU
 
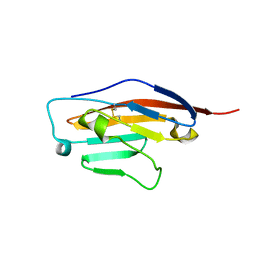 | | Structure of Ig-like V-type domian of mouse Programmed cell death 1 ligand 1 (PD-L1) | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Magiera-Mularz, K, Sala, D, Grudnik, P, Holak, T.A. | | Deposit date: | 2019-09-06 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.532 Å) | | Cite: | Human and mouse PD-L1: similar molecular structure, but different druggability profiles.
Iscience, 24, 2021
|
|
6U6V
 
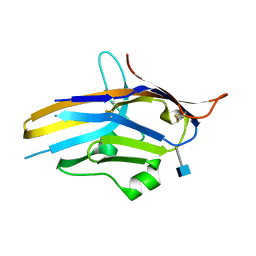 | | Crystal structure of human PD-1H / VISTA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, V-type immunoglobulin domain-containing suppressor of T-cell activation | | Authors: | Slater, B.T, Han, X, Chen, L, Xiong, Y. | | Deposit date: | 2019-08-30 | | Release date: | 2020-01-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insight into T cell coinhibition by PD-1H (VISTA).
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6U3I
 
 | |
6SM1
 
 | | Wild type immunoglobulin light chain (WT-1) | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Immunoglobulin lambda variable 2-14, ... | | Authors: | Kazman, P, Vielberg, M.-T, Cendales, M.D.P, Hunziger, L, Weber, B, Hegenbart, U, Zacharias, M, Koehler, R, Schoenland, S, Groll, M, Buchner, J. | | Deposit date: | 2019-08-21 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fatal amyloid formation in a patient's antibody light chain is caused by a single point mutation.
Elife, 9, 2020
|
|
