4XHD
 
 | | STRUCTURE OF HUMAN PREGNANE X RECEPTOR LIGAND BINDING DOMAIN WITH COMPOUND-1 | | Descriptor: | GLYCEROL, N-{(2R)-1-[(4S)-4-(4-chlorophenyl)-4-hydroxy-3,3-dimethylpiperidin-1-yl]-3-methyl-1-oxobutan-2-yl}-2-cyclopropylacetamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Khan, J.A, Camac, D.M. | | Deposit date: | 2015-01-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Developing Adnectins That Target SRC Co-Activator Binding to PXR: A Structural Approach toward Understanding Promiscuity of PXR.
J.Mol.Biol., 427, 2015
|
|
3WPP
 
 | | Acinetobacter sp. Tol 5 AtaA YDD-DALL3 domains in C-terminal stalk fused to GCN4 adaptors (CstalkC1iii) | | Descriptor: | CHLORIDE ION, Trimeric autotransporter adhesin | | Authors: | Koiwai, K, Hartmann, M.D, Yoshimoto, S, Nur 'Izzah, N, Suzuki, A, Linke, D, Lupas, A.N, Hori, K. | | Deposit date: | 2014-01-15 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structural Basis for Toughness and Flexibility in the C-terminal Passenger Domain of an Acinetobacter Trimeric Autotransporter Adhesin.
J.Biol.Chem., 291, 2016
|
|
1U3Z
 
 | | Crystal structure of MLAC mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
3WQA
 
 | | Acinetobacter sp. Tol 5 AtaA YDD-DALL3 domains in C-terminal stalk fused to GCN4 adaptors (CstalkC1ii) | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Koiwai, K, Hartmann, M.D, Yoshimoto, S, Nur 'Izzah, N, Suzuki, A, Linke, D, Lupas, A.N, Hori, K. | | Deposit date: | 2014-01-24 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural Basis for Toughness and Flexibility in the C-terminal Passenger Domain of an Acinetobacter Trimeric Autotransporter Adhesin.
J.Biol.Chem., 291, 2016
|
|
4XLD
 
 | | Crystal structure of the human PPARg-LBD/rosiglitazone complex obtained by dry co-crystallization and in situ diffraction | | Descriptor: | 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), FORMIC ACID, ... | | Authors: | Delfosse, V, Guichou, J.-F. | | Deposit date: | 2015-01-13 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Combining `dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4Y30
 
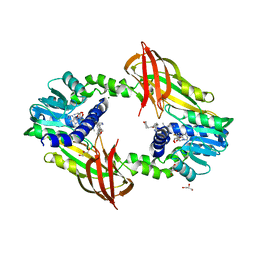 | | Crystal structure of human protein arginine methyltransferase PRMT6 bound to SAH and EPZ020411 | | Descriptor: | GLYCEROL, MAGNESIUM ION, N,N'-dimethyl-N-({3-[4-({trans-3-[2-(tetrahydro-2H-pyran-4-yl)ethoxy]cyclobutyl}oxy)phenyl]-1H-pyrazol-4-yl}methyl)ethane-1,2-diamine, ... | | Authors: | Swinger, K.K, Boriack-Sjodin, P.A. | | Deposit date: | 2015-02-10 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aryl Pyrazoles as Potent Inhibitors of Arginine Methyltransferases: Identification of the First PRMT6 Tool Compound.
Acs Med.Chem.Lett., 6, 2015
|
|
6OJN
 
 | | Comparative Model of SGIV Major Coat Protein (MCP) Trimer Based on Cryo-EM Map | | Descriptor: | Major capsid protein | | Authors: | Pintilie, G, Chen, D.-H, Tran, B.N, Jakana, J, Wu, J, Hew, C.L, Chiu, W. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Segmentation and Comparative Modeling in an 8.6- angstrom Cryo-EM Map of the Singapore Grouper Iridovirus.
Structure, 27, 2019
|
|
4ERZ
 
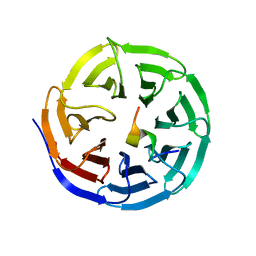 | | X-ray structure of WDR5-MLL4 Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase MLL4, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-21 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
6HWU
 
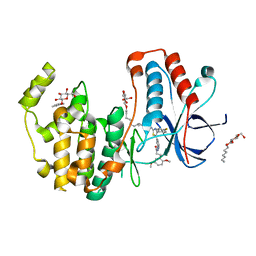 | | Crystal structure of p38alpha in complex with a photoswitchable 2-Azothiazol-based Inhibitor (compound 2) | | Descriptor: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[4-(4-fluorophenyl)-2-[(~{E})-phenyldiazenyl]-1,3-thiazol-5-yl]pyridin-2-yl]propanamide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Mueller, M.P, Rauh, D. | | Deposit date: | 2018-10-15 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2-Azo-, 2-diazocine-thiazols and 2-azo-imidazoles as photoswitchable kinase inhibitors: limitations and pitfalls of the photoswitchable inhibitor approach.
Photochem. Photobiol. Sci., 18, 2019
|
|
3WPR
 
 | | Acinetobacter sp. Tol 5 AtaA N-terminal half of C-terminal stalk fused to GCN4 adaptors (CstalkN) | | Descriptor: | Trimeric autotransporter adhesin | | Authors: | Koiwai, K, Hartmann, M.D, Yoshimoto, S, Nur 'Izzah, N, Suzuki, A, Linke, D, Lupas, A.N, Hori, K. | | Deposit date: | 2014-01-15 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural Basis for Toughness and Flexibility in the C-terminal Passenger Domain of an Acinetobacter Trimeric Autotransporter Adhesin.
J.Biol.Chem., 291, 2016
|
|
3WWT
 
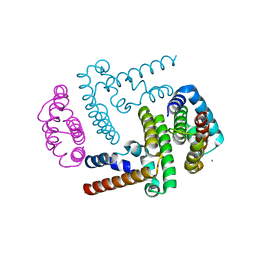 | | Crystal Structure of the Y3:STAT1ND complex | | Descriptor: | C' protein, CALCIUM ION, Signal transducer and activator of transcription 1-alpha/beta | | Authors: | Oda, K, Sakaguchi, T, Matoba, Y. | | Deposit date: | 2014-06-27 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of the Inhibition of STAT1 Activity by Sendai Virus C Protein.
J.Virol., 89, 2015
|
|
7ZJS
 
 | |
4NYC
 
 | | Crystal structure of the E. coli thiM riboswitch in complex with thieno[2,3-b]pyrazin-7-amine | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, thiM TPP riboswitch, ... | | Authors: | Warner, K.D, Homan, P, Weeks, K.M, Smith, A.G, Abell, C, Ferre-D'Amare, A.R. | | Deposit date: | 2013-12-10 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Validating Fragment-Based Drug Discovery for Biological RNAs: Lead Fragments Bind and Remodel the TPP Riboswitch Specifically.
Chem.Biol., 21, 2014
|
|
5IXK
 
 | | RORgamma in complex with inverse agonist BIO399. | | Descriptor: | N-(5-ethyl-3,3-dimethyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-8-yl)-3,4-dimethyl-N-(2,2,2-trifluoroethyl)benzene-1-sulfonamide, Nuclear receptor ROR-gamma | | Authors: | Marcotte, D.J. | | Deposit date: | 2016-03-23 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural determinant for inducing RORgamma specific inverse agonism triggered by a synthetic benzoxazinone ligand.
Bmc Struct.Biol., 16, 2016
|
|
6DMP
 
 | | De Novo Design of a Protein Heterodimer with Specificity Mediated by Hydrogen Bond Networks | | Descriptor: | Designed orthogonal protein DHD13_XAAA_A, Designed orthogonal protein DHD13_XAAA_B | | Authors: | Chen, Z, Flores-Solis, D, Sgourakis, N.G, Baker, D. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
4ERQ
 
 | | X-ray structure of WDR5-MLL2 Win motif peptide binary complex | | Descriptor: | Histone-lysine N-methyltransferase MLL2, WD repeat-containing protein 5 | | Authors: | Dharmarajan, V, Lee, J.-H, Patel, A, Skalnik, D.G, Cosgrove, M.S. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases.
J.Biol.Chem., 287, 2012
|
|
5KI6
 
 | |
5JWQ
 
 | | Crystal structure of KaiC S431E in complex with foldswitch-stabilized KaiB from Thermosynechococcus elongatus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Circadian clock protein KaiB, Circadian clock protein kinase KaiC | | Authors: | Tseng, R, Goularte, N.F, Chavan, A, Luu, J, Chang, Y, Heilser, J, Tripathi, S, LiWang, A, Partch, C.L. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.871 Å) | | Cite: | Structural basis of the day-night transition in a bacterial circadian clock.
Science, 355, 2017
|
|
5K1A
 
 | |
5K3N
 
 | | Crystal structure of Retinoic acid receptor-related orphan receptor (ROR) gamma ligand binding domain complex with ML209 | | Descriptor: | (3S)-3-(2H-1,3-benzodioxol-5-yl)-1-[(3R,5S)-3,5-dimethylpiperidin-1-yl]-3-(2-hydroxy-4,6-dimethoxyphenyl)propan-1-one, Nuclear receptor ROR-gamma | | Authors: | Huang, P, Rastinejad, F, Lu, J. | | Deposit date: | 2016-05-19 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.666 Å) | | Cite: | Crystal structure of Retinoic acid receptor-related orphan receptor (ROR) gamma ligand binding domain complex with ML209
To be published
|
|
4XMN
 
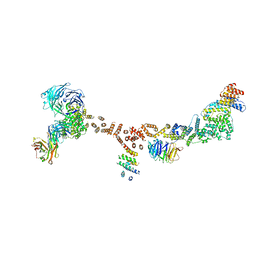 | | Structure of the yeast coat nucleoporin complex, space group P212121 | | Descriptor: | Antibody 87 heavy chain, Antibody 87 light chain, Nucleoporin NUP120, ... | | Authors: | Stuwe, T, Correia, A.R, Lin, D.H, Paduch, M, Lu, V.T, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (7.6 Å) | | Cite: | Nuclear pores. Architecture of the nuclear pore complex coat.
Science, 347, 2015
|
|
4XTA
 
 | |
5K13
 
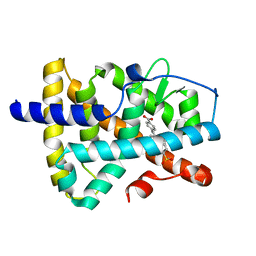 | | Crystal structure of the RAR alpha ligand-binding domain in complex with an antagonist | | Descriptor: | 4-{5-(3-tert-butylphenyl)-1-[4-(methylsulfonyl)phenyl]-1H-pyrazol-3-yl}benzoic acid, Retinoic acid receptor alpha | | Authors: | Wang, Y, Stout, S.L. | | Deposit date: | 2016-05-17 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification of potent and selective retinoic acid receptor gamma (RAR gamma ) antagonists for the treatment of osteoarthritis pain using structure based drug design.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
4Y2H
 
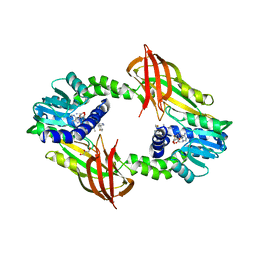 | |
6OCX
 
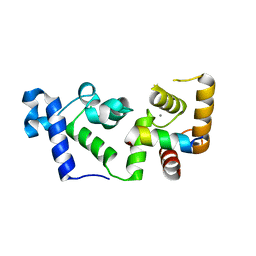 | |
