6Q2X
 
 | |
6DD5
 
 | | Crystal Structure of the Cas6 Domain of Marinomonas mediterranea MMB-1 Cas6-RT-Cas1 Fusion Protein | | Descriptor: | GLYCEROL, MMB-1 Cas6 Fused to Maltose Binding Protein,CRISPR-associated endonuclease Cas1, SULFATE ION, ... | | Authors: | Stamos, J.L, Mohr, G, Silas, S, Makarova, K.S, Markham, L.M, Yao, J, Lucas-Elio, P, Sanchez-Amat, A, Fire, A.Z, Koonin, E.V, Lambowitz, A.M. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Reverse Transcriptase-Cas1 Fusion Protein Contains a Cas6 Domain Required for Both CRISPR RNA Biogenesis and RNA Spacer Acquisition.
Mol. Cell, 72, 2018
|
|
5JNX
 
 | | The 6.6 A cryo-EM structure of the full-length human NPC1 in complex with the cleaved glycoprotein of Ebola virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, ... | | Authors: | Gong, X, Qian, H.W, Zhou, X.H, Wu, J.P, Wan, T, Shi, Y, Gao, F, Zhou, Q, Yan, N. | | Deposit date: | 2016-05-01 | | Release date: | 2016-06-15 | | Last modified: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (6.56 Å) | | Cite: | Structural Insights into the Niemann-Pick C1 (NPC1)-Mediated Cholesterol Transfer and Ebola Infection
Cell, 165, 2016
|
|
7OY7
 
 | | Crystal structure of a trapped Pab-AGOG/double-standed DNA covalent intermediate (DNA containing cytosine opposite to lesion) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*(PED)P*TP*TP*TP*CP*T)-3'), ... | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
6Q9S
 
 | | HDMX (14-111; C17S) COMPLEXED WITH COMPOUND 14 AT 2.4A: Structural states of Hdm2 and HdmX: X-ray elucidation of adaptations and binding interactions for different chemical compound classes | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 3-[[6-chloranyl-3-[3-[(1~{S})-1-(2,4-dichlorophenyl)ethyl]-5-phenyl-imidazol-4-yl]-1~{H}-indol-2-yl]carbonylamino]-4-[4-(2-oxidanylidene-1,3-oxazinan-3-yl)piperidin-1-yl]benzoic acid, Protein Mdm4, ... | | Authors: | Kallen, J. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural States of Hdm2 and HdmX: X-ray Elucidation of Adaptations and Binding Interactions for Different Chemical Compound Classes.
Chemmedchem, 14, 2019
|
|
7AB2
 
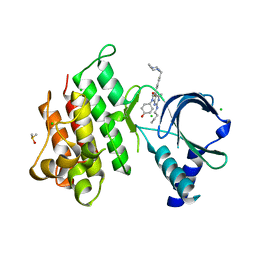 | | Crystal structure of MerTK kinase domain in complex with UNC2025 | | Descriptor: | 4-[2-(butylamino)-5-[4-[(4-methylpiperazin-1-yl)methyl]phenyl]pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexan-1-ol, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Overman, R.C, Rawlins, P.B, Truman, C, Underwood, E, Warwicker, J, Winter-Holt, J. | | Deposit date: | 2020-09-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding.
Biochem.J., 477, 2020
|
|
6YXI
 
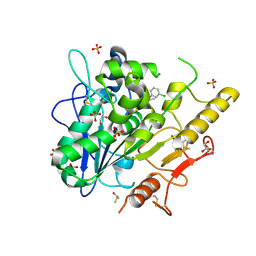 | | Structure of Notum in complex with a 1-(3-Chlorophenyl)-2,5-dimethyl-1H-pyrrole-3-carboxylic acid inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-chlorophenyl)-2,5-dimethyl-pyrrole-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vecchia, L, Jones, E.Y, Ruza, R.R, Hillier, J, Zhao, Y. | | Deposit date: | 2020-05-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6S0H
 
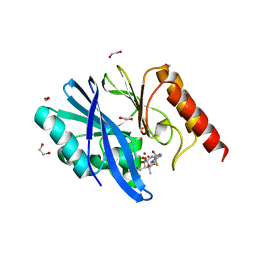 | | Structure of IMP-13 metallo-beta-lactamase complexed with hydrolysed doripenem | | Descriptor: | (2~{R},3~{R})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-3-methyl-4-[(3~{S},5~{S})-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl]sulfanyl-2,3-dihydro-1~{H}-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Zak, K.M, Softley, C, Kolonko, M, Sattler, M, Popowicz, G.M. | | Deposit date: | 2019-06-14 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
7AB1
 
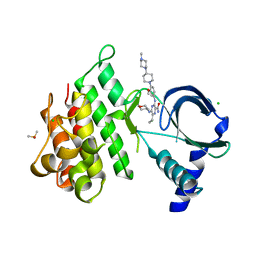 | | Crystal structure of MerTK kinase domain in complex with Gilteritinib | | Descriptor: | 6-ethyl-3-[[3-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]amino]-5-(oxan-4-ylamino)pyrazine-2-carboxamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Overman, R.C, Rawlins, P.B, Truman, C, Underwood, E, Warwicker, J, Winter-Holt, J. | | Deposit date: | 2020-09-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding.
Biochem.J., 477, 2020
|
|
6PV4
 
 | | Structure of CpGH84A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glycoside Hydrolase | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2019-07-19 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of four family 84 glycoside hydrolases from the opportunistic pathogen Clostridium perfringens.
Glycobiology, 30, 2019
|
|
6O1Y
 
 | | Structure of pCW3 conjugation coupling protein TcpA monomeric form with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA translocase coupling protein, beta-D-glucopyranose | | Authors: | Traore, D.A.K, Ahktar, N, Torres, V.T, Adams, V, Coulibaly, F, Panjikar, S, Caradoc-Davies, T.T, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2019-02-22 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of pCW3 conjugation coupling protein TcpA
To be published
|
|
7O4H
 
 | |
1Z4Z
 
 | | Parainfluenza Virus 5 (SV5) Hemagglutinin-Neuraminidase (HN) with ligand DANA(soaked with sialic acid, pH7.0)) | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yuan, P, Thompson, T.B, Wurzburg, B.A, Paterson, R.G, Lamb, R.A, Jardetzky, T.S. | | Deposit date: | 2005-03-16 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose.
Structure, 13, 2005
|
|
4GTW
 
 | | Crystal structure of mouse Enpp1 in complex with AMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kato, K, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-29 | | Release date: | 2012-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Enpp1, an extracellular glycoprotein involved in bone mineralization and insulin signaling.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6Q96
 
 | |
7U2H
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Gly-NH-tRNAgly, aminoacylated P-site fMet-NH-tRNAmet, and deacylated E-site tRNAgly at 2.55A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Syroegin, E.A, Aleksandrova, E.V, Polikanov, Y.S. | | Deposit date: | 2022-02-24 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for the inability of chloramphenicol to inhibit peptide bond formation in the presence of A-site glycine.
Nucleic Acids Res., 50, 2022
|
|
5EZI
 
 | | Crystal Structure of Fab of parasite invasion inhibitory antibody c1 - hexagonal form | | Descriptor: | CHLORIDE ION, Fab c12 Light chain, Fab c12 heavy chain, ... | | Authors: | Favuzza, P, Pluschke, G, Rudolph, M.G. | | Deposit date: | 2015-11-26 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure of the malaria vaccine candidate antigen CyRPA and its complex with a parasite invasion inhibitory antibody.
Elife, 6, 2017
|
|
6QFG
 
 | | Crystal Structure of Human Kallikrein 6 (I218Y) in complex with GSK144 | | Descriptor: | 4-[(5-phenyl-1~{H}-imidazol-2-yl)methylamino]-2-(pyridin-3-ylmethoxy)benzenecarboximidamide, GLYCEROL, Kallikrein-6 | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Evaluation of a crystallographic surrogate for kallikrein 5 in the discovery of novel inhibitors for Netherton syndrome.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
7RBW
 
 | | Structure of Biliverdin-binding Serpin of Boana punctata (polka-dot tree frog) | | Descriptor: | BILIVERDINE IX ALPHA, Biliverdin bindin serpin | | Authors: | Fedorov, E, Manoilov, K.Y, Verkhusha, V, Almo, S.C, Ghosh, A. | | Deposit date: | 2021-07-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Functional Characterization of a Biliverdin-Binding Near-Infrared Fluorescent Protein From the Serpin Superfamily.
J.Mol.Biol., 434, 2021
|
|
1E5J
 
 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN THE TETRAGONAL CRYSTAL FORM IN COMPLEX WITH METHYL-4II-S-ALPHA-CELLOBIOSYL-4II-THIO-BETA-CELLOBIOSIDE | | Descriptor: | CALCIUM ION, ENDOGLUCANASE 5A, alpha-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-methyl beta-D-glucopyranoside | | Authors: | Fort, S, Varrot, A, Schulein, M, Cottaz, S, Driguez, H, Davies, G.J. | | Deposit date: | 2000-07-26 | | Release date: | 2001-07-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mixed-Linkage Cellooligosaccharides: A New Class of Glycoside Hydrolase Inhibitors
Chembiochem, 2, 2001
|
|
4RWF
 
 | |
6Z0K
 
 | | Crystal structure of laccase from Pediococcus acidilactici Pp5930 (Hepes pH 7.5) | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, Putative multicopper oxidase mco | | Authors: | Casino, P, Huesa, J, Pardo, I. | | Deposit date: | 2020-05-09 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis and biochemical properties of laccase enzymes from two Pediococcus species.
Microb Biotechnol, 14, 2021
|
|
6PZ5
 
 | | Crystal Structure of HLA-B*2703 in complex with LRN, a self-peptide | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B*27:03 alpha chain, ... | | Authors: | Gras, S. | | Deposit date: | 2019-07-31 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Allelic association with ankylosing spondylitis fails to correlate with human leukocyte antigen B27 homodimer formation.
J.Biol.Chem., 294, 2019
|
|
3HV3
 
 | | Human p38 MAP Kinase in Complex with RL49 | | Descriptor: | 1-{4-[(6-aminoquinolin-4-yl)amino]phenyl}-3-[3-tert-butyl-1-(3-methylphenyl)-1H-pyrazol-5-yl]urea, GLYCEROL, Mitogen-activated protein kinase 14, ... | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2009-06-15 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Displacement assay for the detection of stabilizers of inactive kinase conformations.
J.Med.Chem., 53, 2010
|
|
7RMW
 
 | | Crystal structure of B. subtilis PurR bound to ppGpp | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, Pur operon repressor | | Authors: | Schumacher, M.A. | | Deposit date: | 2021-07-28 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The nucleotide messenger (p)ppGpp is an anti-inducer of the purine synthesis transcription regulator PurR in Bacillus.
Nucleic Acids Res., 50, 2022
|
|
