6GH0
 
 | |
2JZQ
 
 | | Design of an Active Ultra-Stable Single-Chain Insulin Analog 20 Structures | | Descriptor: | Insulin | | Authors: | Hua, Q.X, Nakarawa, S, Jia, W.H, Huang, K, Philips, N.F, Hu, S.Q, Weiss, M.A. | | Deposit date: | 2008-01-11 | | Release date: | 2008-02-26 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Design of an active ultrastable single-chain insulin analog: synthesis, structure, and therapeutic implications.
J.Biol.Chem., 283, 2008
|
|
2JXZ
 
 | | Solution Conformation of A Non-Amyloidogenic Analogue of Human Calcitonin in Sodium Dodecyl Sulfate Micelles | | Descriptor: | Calcitonin | | Authors: | Andreotti, G, Vitale, R, Avidan-Shpalter, C, Amodeo, P, Gazit, E, Motta, A. | | Deposit date: | 2007-12-03 | | Release date: | 2008-11-25 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Converting the highly amyloidogenic human calcitonin into a powerful fibril inhibitor by 3D structure homology with a non-amyloidogenic analogue
J.Biol.Chem., 286, 2011
|
|
2K1X
 
 | | NMR solution structure of M-crystallin in calcium free form (apo). | | Descriptor: | Beta/gama crystallin family protein | | Authors: | Barnwal, R, Jobby, M, Devi, K, Sharma, Y, Chary, K. | | Deposit date: | 2008-03-17 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and calcium-binding properties of M-crystallin, a primordial betagamma-crystallin from archaea.
J.Mol.Biol., 386, 2009
|
|
2K0Z
 
 | | Solution NMR structure of protein hp1203 from Helicobacter pylori 26695. Northeast Structural Genomics Consortium (NESG) target PT1/Ontario Center for Structural Proteomics target hp1203 | | Descriptor: | Uncharacterized protein hp1203 | | Authors: | Wu, B, Yee, A, Lemak, A, Cort, J, Semest, A, Kenney, M.A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-02-18 | | Release date: | 2008-03-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein hp1203 from Helicobacter pylori 26695. Northeast Structural Genomics Consortium (NESG) target PT1/Ontario Center for Structural Proteomics target hp1203.
To be Published
|
|
2K3M
 
 | | Rv1761c | | Descriptor: | Rv1761c, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Page, R.C, Moore, J.D, Lee, S, Opella, S.J, Cross, T.A. | | Deposit date: | 2008-05-14 | | Release date: | 2009-01-06 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Backbone structure of a small helical integral membrane protein: A unique structural characterization.
Protein Sci., 18, 2009
|
|
2JYO
 
 | | NMR Solution structure of Human MIP-3alpha/CCL20 | | Descriptor: | C-C motif chemokine 20 (Small-inducible cytokine A20) (Macrophage inflammatory protein 3 alpha) (MIP-3-alpha) (Liver and activation-regulated chemokine) (CC chemokine LARC) (Beta chemokine exodus-1) | | Authors: | Chan, D.I, Hunter, H.N, Tack, B.F, Vogel, H.J. | | Deposit date: | 2007-12-14 | | Release date: | 2008-01-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Human macrophage inflammatory protein 3alpha: protein and peptide nuclear magnetic resonance solution structures, dimerization, dynamics, and anti-infective properties.
Antimicrob.Agents Chemother., 52, 2008
|
|
2NDB
 
 | |
2NOZ
 
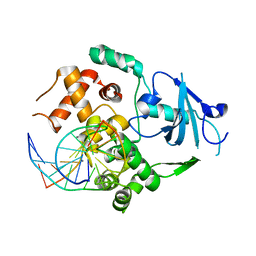 | | Structure of Q315F human 8-oxoguanine glycosylase distal crosslink to 8-oxoguanine DNA | | Descriptor: | 5'-D(*G*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*G*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NOA
 
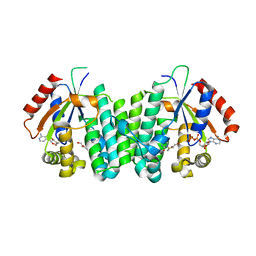 | | The structure of deoxycytidine kinase complexed with lamivudine and ADP. | | Descriptor: | 4-AMINO-1-[(2R,5S)-2-(HYDROXYMETHYL)-1,3-OXATHIOLAN-5-YL]PYRIMIDIN-2(1H)-ONE, ADENOSINE-5'-DIPHOSPHATE, deoxycytidine kinase | | Authors: | Sabini, E, Lavie, A. | | Deposit date: | 2006-10-25 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for activation of the therapeutic L-nucleoside analogs 3TC and troxacitabine by human deoxycytidine kinase.
Nucleic Acids Res., 35, 2007
|
|
2NP2
 
 | | Hbb-DNA complex | | Descriptor: | 36-MER, Hbb | | Authors: | Rice, P.A, Mouw, K.W. | | Deposit date: | 2006-10-26 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Shaping the Borrelia burgdorferi genome: crystal structure and binding properties of the DNA-bending protein Hbb.
Mol.Microbiol., 63, 2007
|
|
2N6M
 
 | |
2NAT
 
 | |
2NAM
 
 | | Full-length WT SOD1 in DPC MICELLE | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Lim, L, Song, J. | | Deposit date: | 2016-01-06 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | SALS-linked WT-SOD1 adopts a highly similar helical conformation as FALS-causing L126Z-SOD1 in a membrane environment
Biochim.Biophys.Acta, 1858, 2016
|
|
2NAU
 
 | |
2NCZ
 
 | |
2NIP
 
 | | NITROGENASE IRON PROTEIN FROM AZOTOBACTER VINELANDII | | Descriptor: | IRON/SULFUR CLUSTER, NITROGENASE IRON PROTEIN | | Authors: | Komiya, H, Georgiadis, M.M, Chakrabarti, P, Woo, D, Kornuc, J.J, Rees, D.C. | | Deposit date: | 1998-05-11 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational variability in structures of the nitrogenase iron proteins from Azotobacter vinelandii and Clostridium pasteurianum.
J.Mol.Biol., 280, 1998
|
|
2NOF
 
 | | Structure of Q315F human 8-oxoguanine glycosylase proximal crosslink to 8-oxoguanine DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*C*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NP3
 
 | | Crystal structure of TetR-family regulator (SCO0857) from Streptomyces coelicolor A3. | | Descriptor: | Putative TetR-family regulator | | Authors: | Koclega, K.D, Xu, X, Chruszcz, M, Gu, J, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of TetR-family regulator (SCO0857) from Streptomyces coelicolor A3.
To be Published
|
|
4KVC
 
 | | 2H2 Fab fragment of immature Dengue virus | | Descriptor: | Ig heavy chain V region MOPC 21, Igh protein, Ig kappa chain V-V region MOPC 21, ... | | Authors: | Wang, Z, Rossmann, M.G. | | Deposit date: | 2013-05-22 | | Release date: | 2013-07-24 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Obstruction of Dengue Virus Maturation by Fab Fragments of the 2H2 Antibody.
J.Virol., 87, 2013
|
|
2NO0
 
 | | C4S dCK variant of dCK in complex with gemcitabine+ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Deoxycytidine kinase, GEMCITABINE | | Authors: | Sabini, E, Hazra, S, Konrad, M, Burley, S.K, Lavie, A. | | Deposit date: | 2006-10-24 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nonenantioselectivity Property of Human Deoxycytidine Kinase Explained by Structures of the Enzyme in Complex with l- and d-Nucleosides.
J.Med.Chem., 50, 2007
|
|
2NOB
 
 | | Structure of catalytically inactive H270A human 8-oxoguanine glycosylase crosslinked to 8-oxoguanine DNA | | Descriptor: | 5'-D(*G*CP*GP*TP*CP*CP*AP*(G42)P*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*T*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NO7
 
 | | C4S dCK variant of dCK in complex with L-dC+ADP | | Descriptor: | 4-AMINO-1-(2-DEOXY-BETA-L-ERYTHRO-PENTOFURANOSYL)PYRIMIDIN-2(1H)-ONE, ADENOSINE-5'-DIPHOSPHATE, deoxycytidine kinase | | Authors: | Sabini, E, Hazra, S, Konrad, M, Burley, S.K, Lavie, A. | | Deposit date: | 2006-10-25 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nonenantioselectivity Property of Human Deoxycytidine Kinase Explained by Structures of the Enzyme in Complex with l- and d-Nucleosides.
J.Med.Chem., 50, 2007
|
|
2NOI
 
 | | Structure of G42A human 8-oxoguanine glycosylase crosslinked to undamaged G-containing DNA | | Descriptor: | 5'-D(*GP*CP*GP*TP*C*CP*AP*GP*GP*TP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', CALCIUM ION, ... | | Authors: | Radom, C.T, Banerjee, A, Verdine, G.L. | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations.
J.Biol.Chem., 282, 2007
|
|
2NLR
 
 | |
