5GKV
 
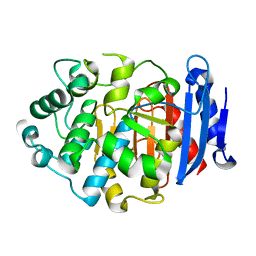 | | Crystal Structure of a Novel Penicillin-Binding Protein (PBP) Homolog from Caulobacter crescentus | | Descriptor: | Esterase A | | Authors: | Ngo, T.D, Ryu, B.H, Kim, B.Y, Yoo, W.K, Lee, E.J, Lee, S.J, Kim, T.D, Kim, K.K. | | Deposit date: | 2016-07-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Biochemical and Structural Analysis of a Novel Penicillin-Binding Protein (PBP) Homolog from Caulobacter crescentus
To Be Published
|
|
6HZI
 
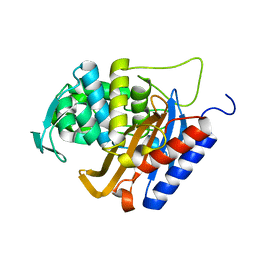 | |
7RD0
 
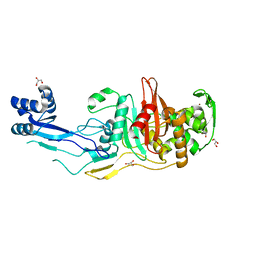 | |
6G0K
 
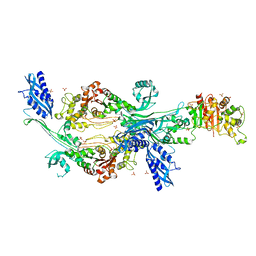 | | Crystal structure of Enterococcus faecium D63r Penicillin-Binding protein 5 (PBP5fm) | | Descriptor: | Low affinity penicillin-binding protein 5 (PBP5), SULFATE ION | | Authors: | Sauvage, E, El Gachi, M, Herman, R, Kerff, F, Charlier, P. | | Deposit date: | 2018-03-19 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of inactivation of Enterococcus faecium penicillin binding protein 5 by ceftobiprole.
To Be Published
|
|
5FQC
 
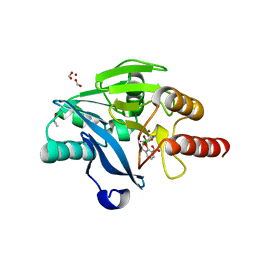 | | Crystal structure of the metallo-beta-lactamase VIM-2 with 2C | | Descriptor: | (4~{R})-4-[[4-(aminomethyl)phenyl]carbonylamino]-3,3-bis(oxidanyl)-2-oxa-3-boranuidabicyclo[4.4.0]deca-1(10),6,8-triene-10-carboxylic acid, BETA-LACTAMASE, DIMETHYL SULFOXIDE, ... | | Authors: | Brem, J, Cain, R, McDonough, M.A, Clifton, I.J, Fishwick, C.W.G, Schofield, C.J. | | Deposit date: | 2015-12-08 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structural basis of metallo-beta-lactamase, serine-beta-lactamase and penicillin-binding protein inhibition by cyclic boronates.
Nat Commun, 7, 2016
|
|
5FQ9
 
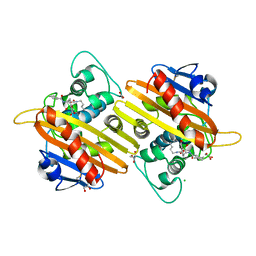 | | Crystal structure of the OXA10 with 1C | | Descriptor: | (3R)-3-(cyclohexylcarbonylamino)-2-oxidanyl-3,4-dihydro-1,2-benzoxaborinine-8-carboxylic acid, ACETATE ION, BETA-LACTAMASE OXA-10, ... | | Authors: | Brem, J, McDonough, M.A, Cain, R, Clifton, I, Fishwick, C.W.G, Schofield, C.J. | | Deposit date: | 2015-12-08 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of metallo-beta-lactamase, serine-beta-lactamase and penicillin-binding protein inhibition by cyclic boronates.
Nat Commun, 7, 2016
|
|
5FQB
 
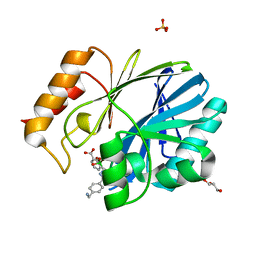 | | Crystal Structure of Bacillus cereus Metallo-Beta-Lactamase with 2C | | Descriptor: | (4~{R})-4-[[4-(aminomethyl)phenyl]carbonylamino]-3,3-bis(oxidanyl)-2-oxa-3-boranuidabicyclo[4.4.0]deca-1(10),6,8-triene-10-carboxylic acid, BETA-LACTAMASE 2, GLYCEROL, ... | | Authors: | Cahill, S.T, Brem, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2015-12-08 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural basis of metallo-beta-lactamase, serine-beta-lactamase and penicillin-binding protein inhibition by cyclic boronates.
Nat Commun, 7, 2016
|
|
3UE1
 
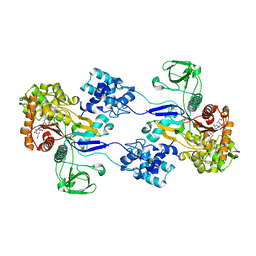 | | Crystal strucuture of Acinetobacter baumanni PBP1A in complex with MC-1 | | Descriptor: | (4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-1-[({4-[(2R)-2,3-dihydroxypropyl]-3-(4,5-dihydroxypyridin-2-yl)-5-oxo-4,5-dihydro- 1H-1,2,4-triazol-1-yl}sulfonyl)amino]-4-formyl-10,10-dimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oate, Penicillin-binding protein 1a | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
3UDI
 
 | |
3UE0
 
 | | Crystal structure of Acinetobacter baumannii PBP1a in complex with Aztreonam | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, Penicillin-binding protein 1a | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
3UDX
 
 | | Crystal structure of Acinetobacter baumannii PBP1a in complex with Imipenem | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 1a | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
1J9M
 
 | | K38H mutant of Streptomyces K15 DD-transpeptidase | | Descriptor: | CHLORIDE ION, DD-transpeptidase, SODIUM ION | | Authors: | Fonze, E, Rhazi, N, Nguyen-Disteche, M, Charlier, P. | | Deposit date: | 2001-05-28 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
2EX2
 
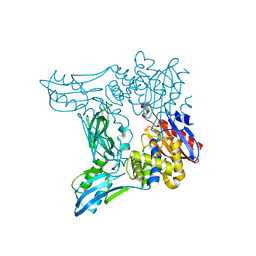 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli | | Descriptor: | GLYCEROL, Penicillin-binding protein 4 | | Authors: | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2005-11-07 | | Release date: | 2006-06-13 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
2EXA
 
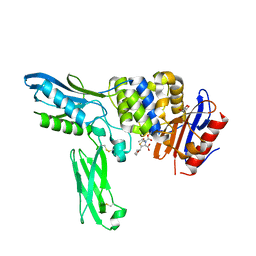 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, complexed with FAROM | | Descriptor: | (2R,5R)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-[(2R)-tetrahydrofuran-2-yl]-2,5-dihydro-1,3-thiazole-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4 | | Authors: | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-06-13 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
2EX8
 
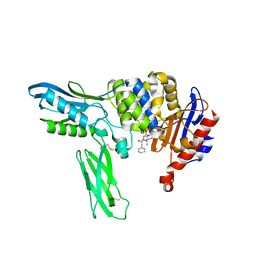 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, complexed with penicillin-G | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin-binding protein 4 | | Authors: | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-06-13 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
2EXB
 
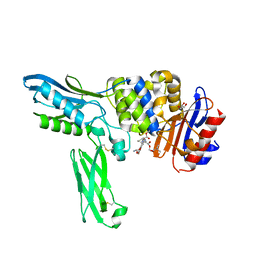 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, complexed with FLOMOX | | Descriptor: | 2,2-dimethylpropanoyloxymethyl (2R)-5-(aminocarbonyloxymethyl)-2-[(1R)-1-[[(Z)-2-(2-azanyl-1,3-thiazol-4-yl)pent-2-enoyl]amino]-2-oxidanylidene-ethyl]-3,6-dihydro-2H-1,3-thiazine-4-carboxylate, GLYCEROL, Penicillin-binding protein 4 | | Authors: | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-06-13 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
2EX9
 
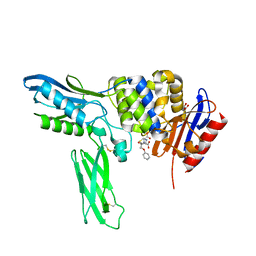 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, complexed with penicillin-V | | Descriptor: | (2R,4S)-5,5-dimethyl-2-{(1R)-2-oxo-1-[(phenoxyacetyl)amino]ethyl}-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4 | | Authors: | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-06-13 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
2EX6
 
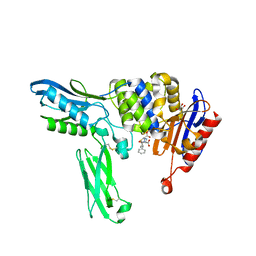 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, complexed with ampicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4 | | Authors: | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-06-13 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
3UDF
 
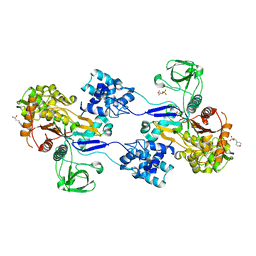 | | Crystal structure of Apo PBP1a from Acinetobacter baumannii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Penicillin-binding protein 1a | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
2FCO
 
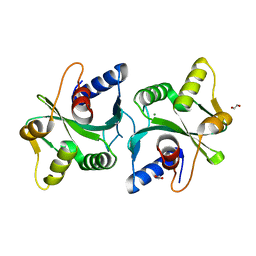 | | Crystal Structure of Bacillus stearothermophilus PrfA-Holliday Junction Resolvase | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, recombination protein U (penicillin-binding protein related factor A) | | Authors: | Li, J, Jedrzejas, M.J. | | Deposit date: | 2005-12-12 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, flexibility, and mechanism of the Bacillus stearothermophilus RecU Holliday junction resolvase.
Proteins, 68, 2007
|
|
3BEC
 
 | |
3BEB
 
 | | Crystal structure of E. coli penicillin-binding protein 5 in complex with a peptide-mimetic penicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(6S)-6-amino-6-carboxyhexanoyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 5 | | Authors: | Heilemann, J, Powell, A.J, Davies, C. | | Deposit date: | 2007-11-16 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of complexes of bacterial DD-peptidases with peptidoglycan-mimetic ligands: the substrate specificity puzzle
J.Mol.Biol., 381, 2008
|
|
1RP5
 
 | | PBP2x from Streptococcus pneumoniae strain 5259 with reduced susceptibility to beta-lactam antibiotics | | Descriptor: | SULFATE ION, penicillin-binding protein 2x | | Authors: | Pernot, L, Chesnel, L, Legouellec, A, Croize, J, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2003-12-03 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A PBP2x from a clinical isolate of Streptococcus pneumoniae exhibits an alternative mechanism for reduction of susceptibility to beta-lactam antibiotics.
J.Biol.Chem., 279, 2004
|
|
2WKE
 
 | | Crystal structure of the Actinomadura R39 DD-peptidase inhibited by 6- beta-iodopenicillanate. | | Descriptor: | (3S)-2,2-dimethyl-3,4-dihydro-2H-1,4-thiazine-3,6-dicarboxylic acid, COBALT (II) ION, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, ... | | Authors: | Sauvage, E, Herman, R, Kerff, F, Charlier, P. | | Deposit date: | 2009-06-10 | | Release date: | 2009-12-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of the Inhibition of Class a Beta-Lactamases and Penicillin-Binding Proteins by 6-Beta-Iodopenicillanate.
J.Am.Chem.Soc., 131, 2009
|
|
2XD5
 
 | | Structural insights into the catalytic mechanism and the role of Streptococcus pneumoniae PBP1b | | Descriptor: | CHLORIDE ION, N-BENZOYL-D-ALANINE, PENICILLIN-BINDING PROTEIN 1B, ... | | Authors: | Macheboeuf, P, Lemaire, D, Jamin, M, Dideberg, O, Dessen, A. | | Deposit date: | 2010-04-29 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism and the Role of Streptococcus Pneumoniae Pbp1B
To be Published
|
|
