3K2M
 
 | |
3KFJ
 
 | |
3M7F
 
 | | Crystal structure of the Nedd4 C2/Grb10 SH2 complex | | Descriptor: | E3 ubiquitin-protein ligase NEDD4, Growth factor receptor-bound protein 10 | | Authors: | Huang, Q, Szebenyi, M. | | Deposit date: | 2010-03-16 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Interaction between the Growth Factor-binding Protein GRB10 and the E3 Ubiquitin Ligase NEDD4.
J.Biol.Chem., 285, 2010
|
|
3MAZ
 
 | | Crystal Structure of the Human BRDG1/STAP-1 SH2 Domain in Complex with the NTAL pTyr136 Peptide | | Descriptor: | CheD family protein, MALONATE ION, Signal-transducing adaptor protein 1 | | Authors: | Kaneko, T, Huang, H, Zhao, B, Li, L, Liu, H, Voss, C.K, Wu, C, Schiller, M.R, Li, S.S. | | Deposit date: | 2010-03-24 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Loops govern SH2 domain specificity by controlling access to binding pockets.
Sci.Signal., 3, 2010
|
|
3MXC
 
 | |
3MXY
 
 | |
3N84
 
 | |
3N7Y
 
 | |
3N8M
 
 | |
3NHN
 
 | | Crystal structure of the SRC-family kinase HCK SH3-SH2-linker regulatory region | | Descriptor: | Tyrosine-protein kinase HCK | | Authors: | Alvarado, J.J, Betts, L, Moroco, J.A, Smithgall, T.E, Yeh, J.I. | | Deposit date: | 2010-06-14 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structure of the Src Family Kinase Hck SH3-SH2 Linker Regulatory Region Supports an SH3-dominant Activation Mechanism.
J.Biol.Chem., 285, 2010
|
|
3OV1
 
 | |
3OVE
 
 | |
2L3T
 
 | | Solution structure of tandem SH2 domain from Spt6 | | Descriptor: | Transcription elongation factor SPT6 | | Authors: | Liu, J, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2010-09-22 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tandem SH2 domains from Spt6 and their binding to the phosphorylated RNA polymerase II C-terminal domain
To be Published
|
|
2L4K
 
 | | Water refined solution structure of the human Grb7-SH2 domain in complex with the 10 amino acid peptide pY1139 | | Descriptor: | Growth factor receptor-bound protein 7, Receptor tyrosine-protein kinase erbB-2 | | Authors: | Pias, S.C, Ivancic, M, Brescia, P.J, Johnson, D.L, Smith, D.E, Daly, R.J, Lyons, B.A. | | Deposit date: | 2010-10-07 | | Release date: | 2010-11-17 | | Last modified: | 2012-10-03 | | Method: | SOLUTION NMR | | Cite: | Water-Refined Solution Structure of the Human Grb7-SH2 Domain in Complex with the erbB2 Receptor Peptide pY1139.
Protein Pept.Lett., 19, 2012
|
|
2L6K
 
 | | Solution Structure of a Nonphosphorylated Peptide Recognizing Domain | | Descriptor: | Tensin-like C1 domain-containing phosphatase | | Authors: | Dai, K, Liao, S, Zhang, J, Zhang, X, Tu, X. | | Deposit date: | 2010-11-22 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Tensin2 SH2 domain and its phosphotyrosine-independent interaction with DLC-1
Plos One, 6, 2011
|
|
3PQZ
 
 | | Grb7 SH2 with peptide | | Descriptor: | Growth factor receptor-bound protein 7, cyclic peptide | | Authors: | Wilce, J.A. | | Deposit date: | 2010-11-29 | | Release date: | 2011-07-20 | | Last modified: | 2011-09-21 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structural basis of binding by cyclic nonphosphorylated Peptide antagonists of grb7 implicated in breast cancer progression
J.Mol.Biol., 412, 2011
|
|
3PS5
 
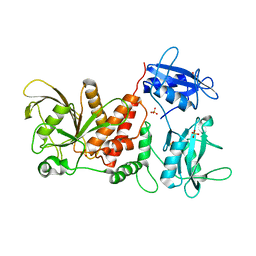 | | Crystal structure of the full-length Human Protein Tyrosine Phosphatase SHP-1 | | Descriptor: | SULFATE ION, Tyrosine-protein phosphatase non-receptor type 6 | | Authors: | Wang, W, Liu, L, Song, X, Mo, Y, Komma, C, Bellamy, H.D, Zhao, Z.J, Zhou, G.W. | | Deposit date: | 2010-11-30 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of human protein tyrosine phosphatase SHP-1 in the open conformation.
J.Cell.Biochem., 112, 2011
|
|
3PSJ
 
 | |
3PSI
 
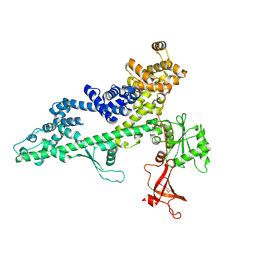 | |
3PSK
 
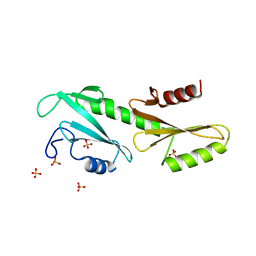 | |
2Y3A
 
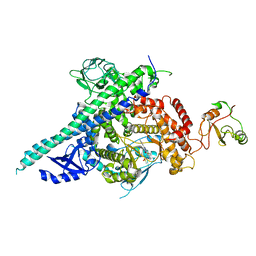 | | Crystal structure of p110beta in complex with icSH2 of p85beta and the drug GDC-0941 | | Descriptor: | 2-(1H-indazol-4-yl)-6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-yl-thieno[3,2-d]pyrimidine, PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY SUBUNIT BETA, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT BETA ISOFORM | | Authors: | Zhang, X, Vadas, O, Perisic, O, Williams, R.L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of Lipid Kinase P110Beta-P85Beta Elucidates an Unusual Sh2-Domain-Mediated Inhibitory Mechanism.
Mol.Cell, 41, 2011
|
|
3QWX
 
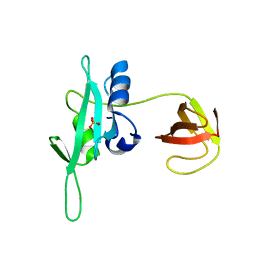 | | CED-2 1-174 | | Descriptor: | Cell death abnormality protein 2, SULFATE ION | | Authors: | Kang, Y, Sun, J, Liu, Y. | | Deposit date: | 2011-02-28 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of the cell corpse engulfment protein CED-2 in Caenorhabditis elegans.
Biochem.Biophys.Res.Commun., 410, 2011
|
|
3QWY
 
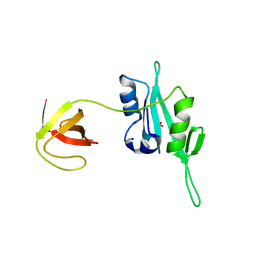 | | CED-2 | | Descriptor: | Cell death abnormality protein 2, GLYCEROL, SULFATE ION | | Authors: | Kang, Y, Sun, J, Liu, Y, Sun, D, Hu, Y, Liu, Y.F. | | Deposit date: | 2011-02-28 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of the cell corpse engulfment protein CED-2 in Caenorhabditis elegans.
Biochem.Biophys.Res.Commun., 410, 2011
|
|
2LCT
 
 | |
3S8N
 
 | |
