6PZD
 
 | |
6PZE
 
 | |
6PZF
 
 | |
2VAB
 
 | |
5UI4
 
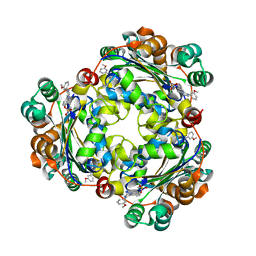 | | Structure of NME1 covalently conjugated to imidazole fluorosulfate | | Descriptor: | 4-[4-(3-methoxyphenyl)-1-(prop-2-yn-1-yl)-1H-imidazol-5-yl]phenyl sulfurofluoridate, Nucleoside diphosphate kinase A | | Authors: | Mortenson, D.E, Brighty, G.J, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2017-01-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | "Inverse Drug Discovery" Strategy To Identify Proteins That Are Targeted by Latent Electrophiles As Exemplified by Aryl Fluorosulfates.
J. Am. Chem. Soc., 140, 2018
|
|
4R26
 
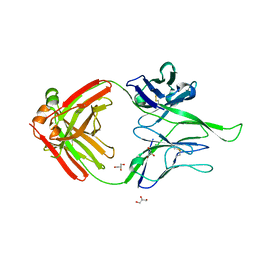 | | Crystal structure of human Fab PGT124, a broadly neutralizing and potent HIV-1 neutralizing antibody | | Descriptor: | GLYCEROL, PGR124-Light Chain, PGT124-Heavy Chain | | Authors: | Garces, F, Kong, L, Wilson, I.A. | | Deposit date: | 2014-08-08 | | Release date: | 2014-10-08 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.4969 Å) | | Cite: | Structural Evolution of Glycan Recognition by a Family of Potent HIV Antibodies.
Cell(Cambridge,Mass.), 159, 2014
|
|
4R2G
 
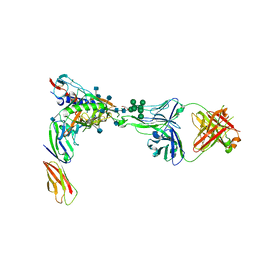 | | Crystal Structure of PGT124 Fab bound to HIV-1 JRCSF gp120 core and to CD4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Garces, F, Wilson, I.A. | | Deposit date: | 2014-08-11 | | Release date: | 2014-10-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | Structural Evolution of Glycan Recognition by a Family of Potent HIV Antibodies.
Cell(Cambridge,Mass.), 159, 2014
|
|
5V2A
 
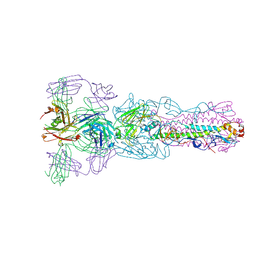 | | Crystal structure of Fab H7.167 in complex with influenza virus hemagglutinin from A/Shanghai/02/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H7.167 antibody, Hemagglutinin, ... | | Authors: | Zhang, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2017-03-03 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.656 Å) | | Cite: | H7N9 influenza virus neutralizing antibodies that possess few somatic mutations.
J. Clin. Invest., 126, 2016
|
|
5KZC
 
 | | Crystal structure of an HIV-1 gp120 engineered outer domain with a Man9 glycan at position N276, in complex with broadly neutralizing antibody VRC01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Engineered outer domain of gp120, VRC01 Fab heavy chain, ... | | Authors: | Julien, J.-P, Jardine, J.G, Diwanji, D, Schief, W.R, Wilson, I.A. | | Deposit date: | 2016-07-24 | | Release date: | 2016-08-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Minimally Mutated HIV-1 Broadly Neutralizing Antibodies to Guide Reductionist Vaccine Design.
Plos Pathog., 12, 2016
|
|
4RBP
 
 | | Crystal structure of HIV neutralizing antibody 2G12 in complex with a bacterial oligosaccharide analog of mammalian oligomanose | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, GLYCEROL, ... | | Authors: | Stanfield, R.L, Wilson, I.A, De Castro, C, Marzaioli, A.M, Pantophlet, R. | | Deposit date: | 2014-09-12 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the HIV neutralizing antibody 2G12 in complex with a bacterial oligosaccharide analog of mammalian oligomannose.
Glycobiology, 25, 2015
|
|
4RQQ
 
 | |
2B1I
 
 | | crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | Bifunctional purine biosynthesis protein PURH, POTASSIUM ION, [3,4-DIHYDROXY-5R-(2,2,4-TRIOXO-1,2R,3S,4R-TETRAHYDRO-2L6-IMIDAZO[4,5-C][1,2,6]THIADIAZIN-7-YL)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN PHOSPHATE | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase.
J.Biol.Chem., 282, 2007
|
|
2B1G
 
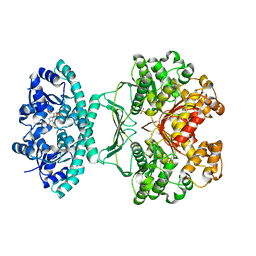 | | Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 7-(3,4-DIHYDROXY-5R-HYDROXYMETHYLTETRAHYDROFURAN-2-YL)-2,2-DIOXO-1,2R,3R,7-TETRAHYDRO-2L6-IMIDAZO[4,5-C][1,2,6]THIADIAZIN-4S-ONE, Bifunctional purine biosynthesis protein PURH, PHOSPHATE ION, ... | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase.
J.Biol.Chem., 282, 2007
|
|
6CJK
 
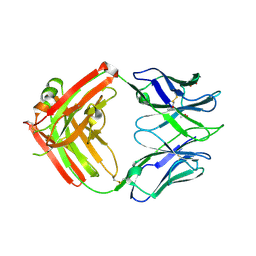 | | Anti HIV Fab 10A | | Descriptor: | ACETATE ION, GLYCEROL, Immunoglobulin Fab heavy chain, ... | | Authors: | Hangartner, L, Ward, A.B, Wilson, I.A, Oyen, D. | | Deposit date: | 2018-02-26 | | Release date: | 2018-08-15 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Electron-Microscopy-Based Epitope Mapping Defines Specificities of Polyclonal Antibodies Elicited during HIV-1 BG505 Envelope Trimer Immunization.
Immunity, 49, 2018
|
|
6CFG
 
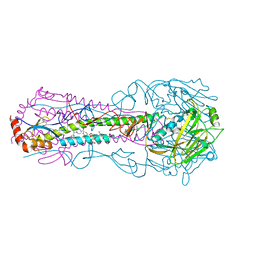 | |
1SXR
 
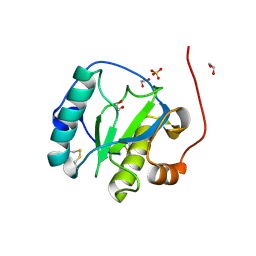 | | Drosophila Peptidoglycan Recognition Protein (PGRP)-SA | | Descriptor: | 1,2-ETHANEDIOL, Peptidoglycan recognition protein SA CG11709-PA, SULFATE ION | | Authors: | Reiser, J.B, Teyton, L, Wilson, I.A. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of the Drosophila peptidoglycan recognition protein (PGRP)-SA at 1.56 A resolution
J.Mol.Biol., 340, 2004
|
|
1TCR
 
 | | MURINE T-CELL ANTIGEN RECEPTOR 2C CLONE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Garcia, K.C, Degano, M, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 1996-09-12 | | Release date: | 1997-03-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex.
Science, 274, 1996
|
|
1T6V
 
 | | Crystal structure analysis of the nurse shark new antigen receptor (NAR) variable domain in complex with lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, novel antigen receptor | | Authors: | Stanfield, R.L, Dooley, H, Flajnik, M.F, Wilson, I.A. | | Deposit date: | 2004-05-07 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a shark single-domain antibody V region in complex with lysozyme.
Science, 305, 2004
|
|
7U2E
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody ADI-55688 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADI-55688 heavy chain, ADI-55688 light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2022-02-23 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A broad and potent neutralization epitope in SARS-related coronaviruses.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7U2D
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody ADG20 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADG20 heavy chain, ADG20 light chain, ... | | Authors: | Zhu, X, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-02-23 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | A broad and potent neutralization epitope in SARS-related coronaviruses.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7T7B
 
 | |
7TP3
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody K288.2 | | Descriptor: | CACODYLATE ION, K288.2 heavy chain, K288.2 light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques.
Sci Transl Med, 14, 2022
|
|
7TP4
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody K398.22 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, K398.22 heavy chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Broadly neutralizing antibodies to SARS-related viruses can be readily induced in rhesus macaques.
Sci Transl Med, 14, 2022
|
|
5E35
 
 | |
5E32
 
 | |
