3MWP
 
 | | Nucleoprotein structure of lassa fever virus | | Descriptor: | Nucleoprotein, ZINC ION | | Authors: | Qi, X, Lan, S, Wang, W, Schelde, L.M, Dong, H, Wallat, G, Liang, Y, Ly, H, Dong, C, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2010-05-06 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Cap binding and immune evasion revealed by Lassa nucleoprotein structure.
Nature, 468, 2010
|
|
3MX5
 
 | | Lassa fever virus nucleoprotein complexed with UTP | | Descriptor: | Nucleoprotein, URIDINE 5'-TRIPHOSPHATE, ZINC ION | | Authors: | Qi, X, Lan, S, Wang, W, Schelde, L.M, Dong, H, Wallat, G, Liang, Y, Ly, H, Dong, C, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2010-05-06 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Cap binding and immune evasion revealed by Lassa nucleoprotein structure.
Nature, 468, 2010
|
|
3MWT
 
 | | Crystal structure of Lassa fever virus nucleoprotein in complex with Mn2+ | | Descriptor: | MANGANESE (II) ION, Nucleoprotein, ZINC ION | | Authors: | Qi, X, Lan, S, Wang, W, Schelde, L.M, Dong, H, Wallat, G, Liang, Y, Ly, H, Dong, C, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2010-05-06 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Cap binding and immune evasion revealed by Lassa nucleoprotein structure.
Nature, 468, 2010
|
|
3MX2
 
 | | Lassa fever virus Nucleoprotein complexed with dTTP | | Descriptor: | Nucleoprotein, THYMIDINE-5'-TRIPHOSPHATE, ZINC ION | | Authors: | Qi, X, Lan, S, Wang, W, Schelde, L.M, Dong, H, Wallat, G, Liang, Y, Ly, H, Dong, C, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2010-05-06 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Cap binding and immune evasion revealed by Lassa nucleoprotein structure.
Nature, 468, 2010
|
|
4KZM
 
 | | Crystal Structure of TR3 LBD S553A Mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Zhang, Q, Li, A, Tian, X, Cai, Q, Wang, W, Wang, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
7V4Y
 
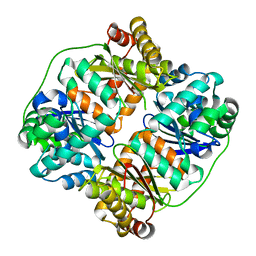 | | TTHA1264/TTHA1265 complex | | Descriptor: | Putative zinc protease, ZINC ION, Zinc-dependent peptidase | | Authors: | Xu, M, Xu, Q, Ran, T, Wang, W, Sun, B, Wang, Q. | | Deposit date: | 2021-08-16 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of TTHA1265 and TTHA1264/TTHA1265 complex reveal an intrinsic heterodimeric assembly.
Int.J.Biol.Macromol., 207, 2022
|
|
6CQD
 
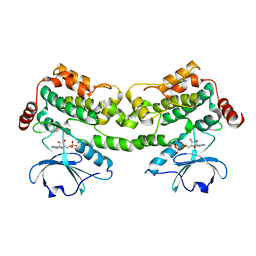 | | Crystal structure of HPK1 in complex with ATP analogue (AMPPNP) | | Descriptor: | MAGNESIUM ION, Mitogen-activated protein kinase kinase kinase kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wu, P, Lehoux, I, Franke, Y, Mortara, K, Wang, W. | | Deposit date: | 2018-03-14 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Hematopoietic Progenitor Kinase-1 Structure in a Domain-Swapped Dimer.
Structure, 27, 2019
|
|
6CQF
 
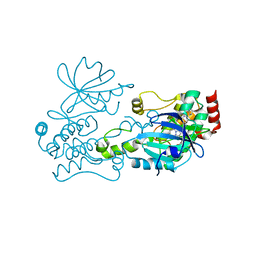 | | Crystal structure of HPK1 in complex an inhibitor G1858 | | Descriptor: | Mitogen-activated protein kinase kinase kinase kinase 1, N-{2-(3,3-difluoropyrrolidin-1-yl)-6-[(3R)-pyrrolidin-3-yl]pyrimidin-4-yl}-1-(propan-2-yl)-1H-pyrazolo[4,3-c]pyridin-6-amine | | Authors: | Wu, P, Lehoux, I, Mortara, K, Franke, Y, Chan, B.K, Wang, W. | | Deposit date: | 2018-03-15 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Hematopoietic Progenitor Kinase-1 Structure in a Domain-Swapped Dimer.
Structure, 27, 2019
|
|
6D5Y
 
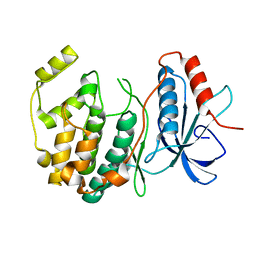 | | Crystal structure of ERK2 G169D mutant | | Descriptor: | Mitogen-activated protein kinase 1 | | Authors: | Yin, J, Jaiswal, B.S, Wang, W. | | Deposit date: | 2018-04-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | ERK Mutations and Amplification Confer Resistance to ERK-Inhibitor Therapy.
Clin. Cancer Res., 24, 2018
|
|
6CQE
 
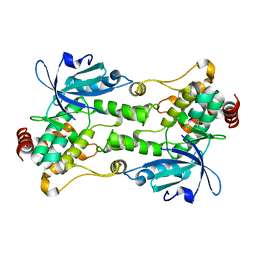 | | Crystal structure of HPK1 kinase domain S171A mutant | | Descriptor: | Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Wu, P, Lehoux, I, Mortara, K, Franke, Y, Wang, W. | | Deposit date: | 2018-03-15 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.886 Å) | | Cite: | Hematopoietic Progenitor Kinase-1 Structure in a Domain-Swapped Dimer.
Structure, 27, 2019
|
|
7E59
 
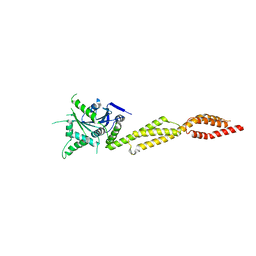 | | interferon-inducible anti-viral protein truncated | | Descriptor: | Guanylate-binding protein 5 | | Authors: | Cui, W, Wang, W, Chen, C, Slater, B, Xiong, Y, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-02-18 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7E58
 
 | | interferon-inducible anti-viral protein 2 | | Descriptor: | Guanylate-binding protein 2 | | Authors: | Cui, W, Wang, W, Chen, C, Slater, B, Xiong, Y, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-02-18 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7E5A
 
 | | interferon-inducible anti-viral protein R356A | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, Guanylate-binding protein 5, ... | | Authors: | Cui, W, Wang, W, Chen, C, Slater, B, Xiong, Y, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-02-18 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EW2
 
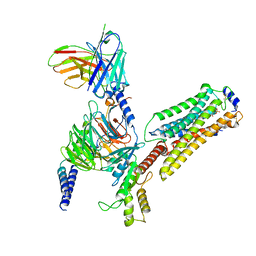 | | Cryo-EM structure of pFTY720-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | Descriptor: | (2~{S})-2-azanyl-4-(4-octylphenyl)-2-[[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxymethyl]butan-1-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
7EW4
 
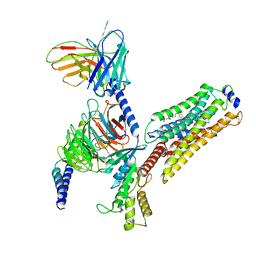 | | Cryo-EM structure of CYM-5541-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
7EW3
 
 | | Cryo-EM structure of S1P-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
6J9R
 
 | |
6J36
 
 | | crystal structure of Mycoplasma hyopneumoniae Enolase | | Descriptor: | Enolase, GLYCEROL, SULFATE ION | | Authors: | Chen, R, Zhang, S, Gan, R, Xie, X, Feng, Z, Wang, W, Ran, T, Zhang, W, Xiang, Q, Shao, G. | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Featured Species-Specific Loops Are Found in the Crystal Structure ofMhpEno, a Cell Surface Adhesin FromMycoplasma hyopneumoniae.
Front Cell Infect Microbiol, 9, 2019
|
|
8WCK
 
 | | FCP tetramer in Chaetoceros gracilis | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, CHLOROPHYLL A, Chlorophyll a/b-binding protein, ... | | Authors: | Feng, Y, Li, Z, Zhou, C, Shen, J.-R, Liu, C, Wang, W. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural and spectroscopic insights into fucoxanthin chlorophyll a/c-binding proteins of diatoms in diverse oligomeric states.
Plant Commun., 2024
|
|
8WCL
 
 | | FCP pentamer in Chaetoceros gracilis | | Descriptor: | (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, CHLOROPHYLL A, ... | | Authors: | Feng, Y, Li, Z, Zhou, C, Liu, C, Shen, J.-R, Wang, W. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural and spectroscopic insights into fucoxanthin chlorophyll a/c-binding proteins of diatoms in diverse oligomeric states.
Plant Commun., 2024
|
|
4I35
 
 | | The crystal structure of serralysin | | Descriptor: | CALCIUM ION, GLYCEROL, HEXANE, ... | | Authors: | Zou, M, Ran, T, Xu, D, Wang, W. | | Deposit date: | 2012-11-24 | | Release date: | 2013-11-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | The crystal structure of serralysin
To be Published
|
|
7VIB
 
 | | Crystal structure of human ACE2 and GX/P2V RBD | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein, ZINC ION | | Authors: | Guo, Y, Cao, W, Jia, N, Wang, W, Yuan, S, Wang, Y. | | Deposit date: | 2021-09-26 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of human ACE2 and GX/P2V RBD
To Be Published
|
|
3ZEE
 
 | | Electron cyro-microscopy helical reconstruction of Par-3 N terminal domain | | Descriptor: | PARTITIONING DEFECTIVE 3 HOMOLOG | | Authors: | Zhang, Y, Wang, W, Chen, J, Zhang, K, Gao, F, Gong, W, Zhang, M, Sun, F, Feng, W. | | Deposit date: | 2012-12-05 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural Insights Into the Intrinsic Self-Assembly of Par-3 N-Terminal Domain.
Structure, 21, 2013
|
|
4O0Y
 
 | | Back pocket flexibility provides group-II PAK selectivity for type 1 kinase inhibitors | | Descriptor: | 4-[1-(4-amino-1,3,5-triazin-2-yl)-2-(ethylamino)-1H-benzimidazol-6-yl]-2-methylbut-3-yn-2-ol, Serine/threonine-protein kinase PAK 4 | | Authors: | Rouge, L, Tam, C, Wang, W. | | Deposit date: | 2013-12-14 | | Release date: | 2014-02-12 | | Last modified: | 2014-02-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Back Pocket Flexibility Provides Group II p21-Activated Kinase (PAK) Selectivity for Type I 1/2 Kinase Inhibitors.
J.Med.Chem., 57, 2014
|
|
4O19
 
 | | The crystal structure of a mutant NAMPT (G217V) | | Descriptor: | 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
