6VRW
 
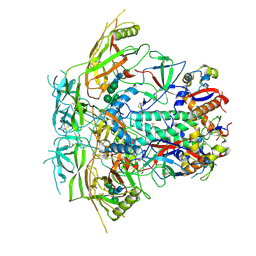 | | Cryo-EM structure of stabilized HIV-1 Env trimer CAP256.wk34.c80 SOSIP.RnS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2020-02-10 | | Release date: | 2020-03-11 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structure of Super-Potent Antibody CAP256-VRC26.25 in Complex with HIV-1 Envelope Reveals a Combined Mode of Trimer-Apex Recognition.
Cell Rep, 31, 2020
|
|
6VRY
 
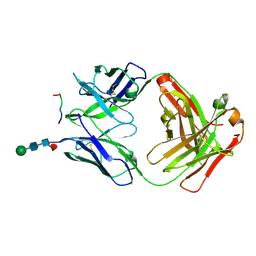 | |
6VZI
 
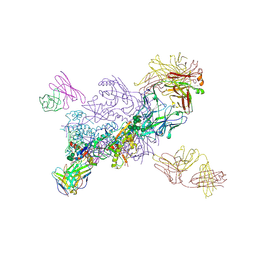 | |
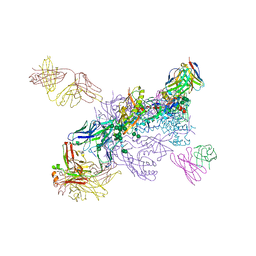
6W03
 
 | |
6VTT
 
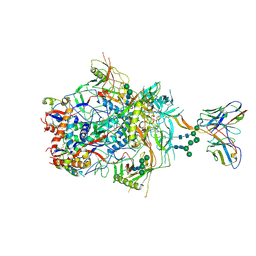 | |
6WIX
 
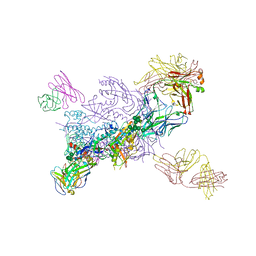 | | Crystal Structure of HIV-1 MI369 RnS-DS.SOSIP Prefusion Env Trimer in Complex with Human Antibodies 3H109L and 35O22 at 3.5 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lai, Y.-T, Olia, A, Kwong, P.D. | | Deposit date: | 2020-04-10 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Automated Design by Structure-Based Stabilization and Consensus Repair to Achieve Prefusion-Closed Envelope Trimers in a Wide Variety of HIV Strains.
Cell Rep, 33, 2020
|
|
5TKJ
 
 | | Structure of vaccine-elicited diverse HIV-1 neutralizing antibody vFP1.01 in complex with HIV-1 fusion peptide residue 512-519 | | Descriptor: | HIV-1 fusion peptide residue 512-519, SULFATE ION, vFP1.01 chimeric mouse antibody heavy chain, ... | | Authors: | Xu, K, Liu, K, Kwong, P.D. | | Deposit date: | 2016-10-06 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.118 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
5TE7
 
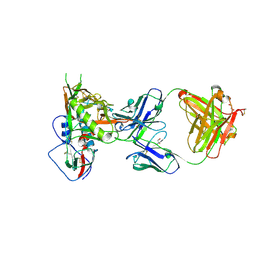 | |
5TY6
 
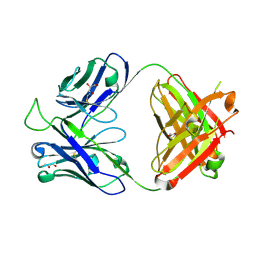 | | Crystal structure of the broadly neutralizing Influenza A antibody VRC 315 13-1b02 Fab. | | Descriptor: | GLYCEROL, VRC 315 13-1b02 Fab Heavy chain, VRC 315 13-1b02 Fab Light chain | | Authors: | Joyce, M.G, Andrews, S.F, Mascola, J.R, McDermott, A.B, Kwong, P.D. | | Deposit date: | 2016-11-18 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.361 Å) | | Cite: | Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans.
Sci Immunol, 2, 2017
|
|
5U1F
 
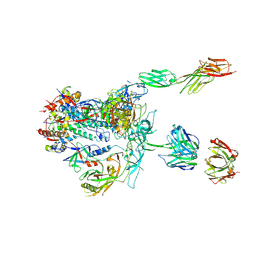 | | Initial contact of HIV-1 Env with CD4: Cryo-EM structure of BG505 DS-SOSIP trimer in complex with CD4 and antibody PGT145 | | Descriptor: | BG505 DS-SOSIP gp120, BG505 SOSIP gp41, PGT145 heavy chain, ... | | Authors: | Acharya, P, Kwong, P.D, Potter, C.S, Carragher, B. | | Deposit date: | 2016-11-28 | | Release date: | 2017-02-22 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Quaternary contact in the initial interaction of CD4 with the HIV-1 envelope trimer.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5U4R
 
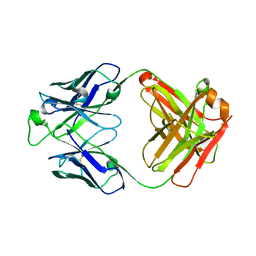 | | Crystal structure of the broadly neutralizing Influenza A antibody VRC 315 53-1A09 Fab. | | Descriptor: | VRC 315 53-1A09 Fab Heavy chain, VRC 315 53-1A09 Fab Light chain | | Authors: | Joyce, M.G, Andrews, S.F, Mascola, J.R, McDermott, A.B, Kwong, P.D. | | Deposit date: | 2016-12-05 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans.
Sci Immunol, 2, 2017
|
|
5TR8
 
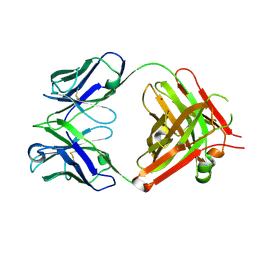 | | Crystal structure of vaccine-elicited pan- influenza H1N1 neutralizing murine antibody 441D6. | | Descriptor: | 441D6 Fab Heavy chain, 441D6 Fab Light chain, NICKEL (II) ION | | Authors: | Joyce, M.G, Kanekiyo, M, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2016-10-25 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses.
Nat.Immunol., 20, 2019
|
|
5U6E
 
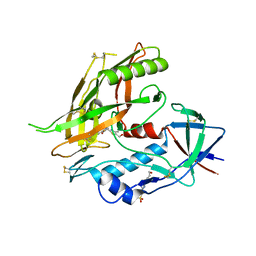 | | Crystal structure of clade A/E HIV-1 gp120 core in complex with NBD-14010 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-{(1S)-2-amino-1-[5-(hydroxymethyl)-4-methyl-1,3-thiazol-2-yl]ethyl}-5-(4-chloro-3-fluorophenyl)-1H-pyrrole-2-carboxamide, ... | | Authors: | Kwon, Y.D, Debnath, A.K, Kwong, P.D. | | Deposit date: | 2016-12-07 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Synthesis, Antiviral Potency, in Vitro ADMET, and X-ray Structure of Potent CD4 Mimics as Entry Inhibitors That Target the Phe43 Cavity of HIV-1 gp120.
J. Med. Chem., 60, 2017
|
|
5U7M
 
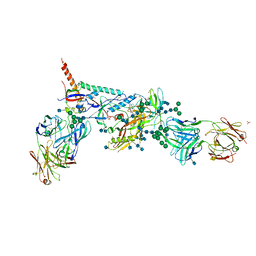 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer Bound to Small Molecule HIV-1 Entry Inhibitor BMS-378806 in Complex with Human Antibodies PGT122 and 35O22 at 3.8 Angstrom | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pancera, M, Lai, Y.-T, Kwong, P.D. | | Deposit date: | 2016-12-12 | | Release date: | 2017-08-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.025 Å) | | Cite: | Crystal structures of trimeric HIV envelope with entry inhibitors BMS-378806 and BMS-626529.
Nat. Chem. Biol., 13, 2017
|
|
5TDG
 
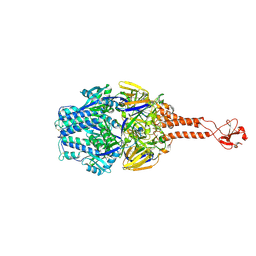 | |
5UTY
 
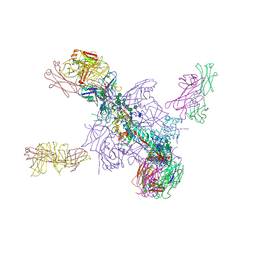 | | Crystal Structure of a Stabilized DS-SOSIP.mut4 BG505 gp140 HIV-1 Env Trimer, Containing Mutations I201C-P433C (DS), L154M, N300M, N302M, T320L in Complex with Human Antibodies PGT122 and 35O22 at 4.1 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Fab heavy chain, ... | | Authors: | Xu, K, Chuang, G.-Y, Pancera, M, Kwong, P.D. | | Deposit date: | 2017-02-15 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.412 Å) | | Cite: | Structure-Based Design of a Soluble Prefusion-Closed HIV-1 Env Trimer with Reduced CD4 Affinity and Improved Immunogenicity.
J. Virol., 91, 2017
|
|
5T33
 
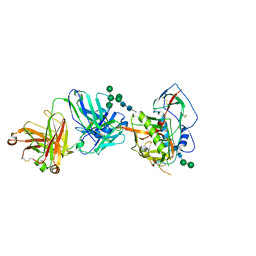 | | Crystal structure of strain-specific glycan-dependent CD4 binding site-directed neutralizing antibody CAP257-RH1, in complex with HIV-1 strain RHPA gp120 core with an oligomannose N276 glycan. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CAP257-RH1 heavy chain, ... | | Authors: | Wibmer, C.K, Gorman, J, Kwong, P.D. | | Deposit date: | 2016-08-24 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2092 Å) | | Cite: | Structure of an N276-Dependent HIV-1 Neutralizing Antibody Targeting a Rare V5 Glycan Hole Adjacent to the CD4 Binding Site.
J.Virol., 90, 2016
|
|
5U7O
 
 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer Bound to Small Molecule HIV-1 Entry Inhibitor BMS-626529 in Complex with Human Antibodies PGT122 and 35O22 at 3.8 Angstrom | | Descriptor: | 1-[4-(benzenecarbonyl)piperazin-1-yl]-2-[4-methoxy-7-(3-methyl-1H-1,2,4-triazol-1-yl)-1H-pyrrolo[2,3-c]pyridin-3-yl]ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pancera, M, Lai, Y.-T, Kwong, P.D. | | Deposit date: | 2016-12-12 | | Release date: | 2017-08-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.031 Å) | | Cite: | Crystal structures of trimeric HIV envelope with entry inhibitors BMS-378806 and BMS-626529.
Nat. Chem. Biol., 13, 2017
|
|
5UTF
 
 | | Crystal Structure of a Stabilized DS-SOSIP.6mut BG505 gp140 HIV-1 Env Trimer, Containing Mutations I201C-P433C (DS), L154M, Y177W, N300M, N302M, T320L, I420M in Complex with Human Antibodies PGT122 and 35O22 at 4.3 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35022 Heavy chain, ... | | Authors: | Pancera, M, Chuang, G.-Y, Xu, K, Kwong, P.D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Structure-Based Design of a Soluble Prefusion-Closed HIV-1 Env Trimer with Reduced CD4 Affinity and Improved Immunogenicity.
J. Virol., 91, 2017
|
|
5V7J
 
 | | Crystal Structure at 3.7 A Resolution of Glycosylated HIV-1 Clade A BG505 SOSIP.664 Prefusion Env Trimer with Four Glycans (N197, N276, N362, and N462) removed in Complex with Neutralizing Antibodies 3H+109L and 35O22. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 35O22 Fab heavy chain, ... | | Authors: | Stewart-Jones, G.B.E, Zhou, T, Kwong, P.D. | | Deposit date: | 2017-03-20 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | Quantification of the Impact of the HIV-1-Glycan Shield on Antibody Elicitation.
Cell Rep, 19, 2017
|
|
5TDL
 
 | |
5TE6
 
 | |
5TKK
 
 | |
5TE4
 
 | | Crystal Structure of Broadly Neutralizing VRC01-class Antibody N6 in Complex with HIV-1 Clade G Strain X2088 gp120 Core | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, T, Kwong, P.D. | | Deposit date: | 2016-09-20 | | Release date: | 2016-11-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification of a CD4-Binding-Site Antibody to HIV that Evolved Near-Pan Neutralization Breadth.
Immunity, 45, 2016
|
|
5VGJ
 
 | | Crystal Structure of the Human Fab VRC38.01, an HIV-1 V1V2-Directed Neutralizing Antibody Isolated from Donor N90, bound to a scaffolded WITO V1V2 domain | | Descriptor: | 1FD6-V1V2-WITO, 2-acetamido-2-deoxy-beta-D-glucopyranose, VRC38.01 Fab Heavy Chain, ... | | Authors: | Gorman, J, Li, J, Kwong, P.D. | | Deposit date: | 2017-04-11 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.456 Å) | | Cite: | Virus-like Particles Identify an HIV V1V2 Apex-Binding Neutralizing Antibody that Lacks a Protruding Loop.
Immunity, 46, 2017
|
|
