1J20
 
 | |
1X28
 
 | | Crystal Structure of e.coli AspAT complexed with N-phosphopyridoxyl-L-glutamic acid | | Descriptor: | Aspartate aminotransferase, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-glutamic acid | | Authors: | Goto, M. | | Deposit date: | 2005-04-21 | | Release date: | 2005-06-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Binding of C5-dicarboxylic substrate to aspartate aminotransferase: implications for the conformational change at the transaldimination step.
Biochemistry, 44, 2005
|
|
1X29
 
 | | Crystal Structure of e.coli AspAT complexed with N-phosphopyridoxyl-2-methyl-L-glutamic acid | | Descriptor: | Aspartate aminotransferase, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-2-METHYL-L-GLUTAMIC ACID | | Authors: | Goto, M. | | Deposit date: | 2005-04-21 | | Release date: | 2005-06-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of C5-dicarboxylic substrate to aspartate aminotransferase: implications for the conformational change at the transaldimination step.
Biochemistry, 44, 2005
|
|
1WRV
 
 | |
4TV7
 
 | |
2A1H
 
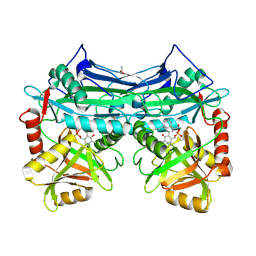 | | X-ray crystal structure of human mitochondrial branched chain aminotransferase (BCATm) complexed with gabapentin | | Descriptor: | ACETIC ACID, PYRIDOXAL-5'-PHOSPHATE, [1-(AMINOMETHYL)CYCLOHEXYL]ACETIC ACID, ... | | Authors: | Goto, M, Miyahara, I, Hirotsu, K, Conway, M, Yennawar, N, Islam, M.M, Hutson, S.M. | | Deposit date: | 2005-06-20 | | Release date: | 2005-09-06 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural determinants for branched-chain aminotransferase isozyme-specific inhibition by the anticonvulsant drug gabapentin.
J.Biol.Chem., 280, 2005
|
|
6LUT
 
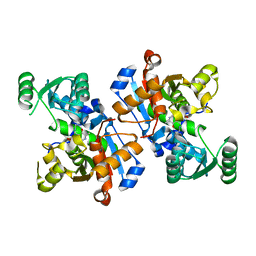 | |
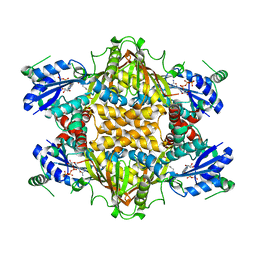
1J21
 
 | |
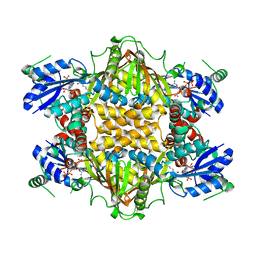
1J1Z
 
 | |
5AVO
 
 | |
1KH3
 
 | | Crystal Structure of Thermus thermophilus HB8 Argininosuccinate Synthetase in complex with inhibitor | | Descriptor: | ARGININE, ASPARTIC ACID, Argininosuccinate Synthetase, ... | | Authors: | goto, m, Hirotsu, k, miyahara, i, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-11-29 | | Release date: | 2003-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Argininosuccinate Synthetase in Enzyme-ATP Substrates and Enzyme-AMP Product Forms: STEREOCHEMISTRY OF THE CATALYTIC REACTION
J.Biol.Chem., 278, 2003
|
|
1KOR
 
 | | Crystal Structure of Thermus thermophilus HB8 Argininosuccinate Synthetase in complex with inhibitors | | Descriptor: | ARGININE, Argininosuccinate Synthetase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Goto, M, Nakajima, Y, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-12-22 | | Release date: | 2002-04-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of argininosuccinate synthetase from Thermus thermophilus HB8. Structural basis for the catalytic action.
J.Biol.Chem., 277, 2002
|
|
1KH2
 
 | |
1KH1
 
 | |
4YDR
 
 | |
5X9D
 
 | | Crystal structure of homoserine dehydrogenase in complex with L-cysteine and NAD | | Descriptor: | (2R)-3-[[(4S)-3-aminocarbonyl-1-[(2R,3R,4S,5R)-5-[[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-3,4-bis(oxidanyl)oxolan-2-yl]-4H-pyridin-4-yl]sulfanyl]-2-azanyl-propanoic acid, Homoserine dehydrogenase, L(+)-TARTARIC ACID | | Authors: | Goto, M, Ogata, K, Kaneko, R, Yoshimune, K. | | Deposit date: | 2017-03-06 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of homoserine dehydrogenase by formation of a cysteine-NAD covalent complex
Sci Rep, 8, 2018
|
|
1X2A
 
 | | Crystal Structure of e.coli AspAT complexed with N-phosphopyridoxyl-D-glutamic acid | | Descriptor: | Aspartate aminotransferase, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-D-GLUTAMIC ACID | | Authors: | Goto, M. | | Deposit date: | 2005-04-21 | | Release date: | 2005-06-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of C5-dicarboxylic substrate to aspartate aminotransferase: implications for the conformational change at the transaldimination step.
Biochemistry, 44, 2005
|
|
8I7P
 
 | | Crystal structure of Ricin A chain bound with N2-(2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)glycyl-L-tyrosine | | Descriptor: | 6-(2-ethyl-4-hydroxyphenyl)-1H-indazole-3-carboxamide, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Sakamoto, N, Higashi, S, Kawata, R, Nagatsu, K, Saito, R. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of ricin toxin A chain complexed with a highly potent pterin-based small-molecular inhibitor.
J Enzyme Inhib Med Chem, 38, 2023
|
|
2COG
 
 | |
2COI
 
 | |
7XZS
 
 | | Crystal structure of Ricin A chain bound with (2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)-L-tyrosine | | Descriptor: | (2S)-2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]-3-(4-hydroxyphenyl)propanoic acid, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
7XZT
 
 | | Crystal structure of Ricin A chain bound with (2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)-D-tyrosine | | Descriptor: | (2R)-2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]-3-(4-hydroxyphenyl)propanoic acid, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
7XZW
 
 | | Crystal structure of Ricin A chain bound with (2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)-D-phenylalanine | | Descriptor: | (2R)-2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]-3-phenyl-propanoic acid, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
7XZU
 
 | | Crystal structure of Ricin A chain bound with (2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)-L-phenylalanine | | Descriptor: | (2S)-2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]-3-phenyl-propanoic acid, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
7Y02
 
 | | Crystal structure of Ricin A chain bound with (S)-2-(2-amino-4-oxo-3,4-dihydropteridine-7-carboxamido)-3-(4-fluorophenyl)propanoic acid | | Descriptor: | (2S)-2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]-3-(4-fluorophenyl)propanoic acid, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Higashi, S, Ohba, T, Kawata, R, Nagatsu, K, Suzuki, S, Saito, R. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational change in ricin toxin A-Chain: A critical factor for inhibitor binding to the secondary pocket.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
