6IX3
 
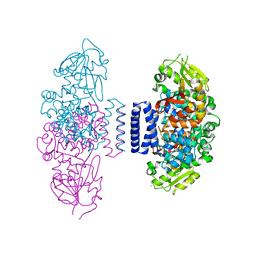 | | The structure of LepI complex with SAM | | Descriptor: | CHLORIDE ION, O-methyltransferase lepI, S-ADENOSYLMETHIONINE | | Authors: | Cai, Y, Ohashi, M, Hai, Y, Tang, Y, Zhou, J. | | Deposit date: | 2018-12-09 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis for stereoselective dehydration and hydrogen-bonding catalysis by the SAM-dependent pericyclase LepI.
Nat.Chem., 11, 2019
|
|
6IX9
 
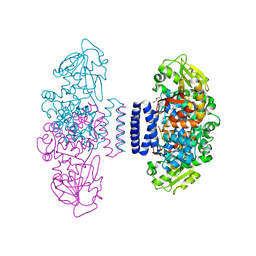 | | The structure of LepI C52A in complex with SAM and leporin C | | Descriptor: | (6R,6aS,10S,10aR)-10-methyl-4-phenyl-6-[(1E)-prop-1-en-1-yl]-2,6,6a,7,8,9,10,10a-octahydro-1H-[2]benzopyrano[4,3-c]pyridin-1-one, CHLORIDE ION, GLYCEROL, ... | | Authors: | Cai, Y, Ohashi, M, Hai, Y, Tang, Y, Zhou, J. | | Deposit date: | 2018-12-09 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Structural basis for stereoselective dehydration and hydrogen-bonding catalysis by the SAM-dependent pericyclase LepI.
Nat.Chem., 11, 2019
|
|
6IX7
 
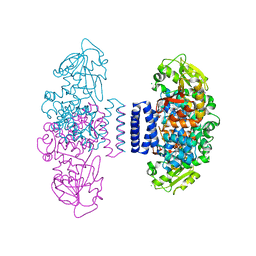 | | The structure of LepI C52A in complex with SAH and substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-3-[(2S,6E,8E)-2-methyldeca-6,8-dienoyl]-5-phenylpyridin-2(1H)-one, CHLORIDE ION, ... | | Authors: | Cai, Y, Ohashi, M, Hai, Y, Tang, Y, Zhou, J. | | Deposit date: | 2018-12-09 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.835 Å) | | Cite: | Structural basis for stereoselective dehydration and hydrogen-bonding catalysis by the SAM-dependent pericyclase LepI.
Nat.Chem., 11, 2019
|
|
6L29
 
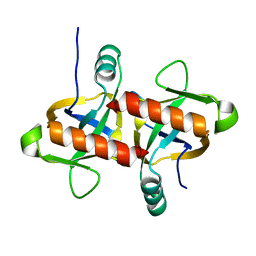 | | The structure of the MazF-mt1 mutant | | Descriptor: | mRNA interferase | | Authors: | Xie, W, Chen, R, Zhou, J. | | Deposit date: | 2019-10-02 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3000052 Å) | | Cite: | Conserved Conformational Changes in the Regulation ofMycobacterium tuberculosisMazEF-mt1.
Acs Infect Dis., 6, 2020
|
|
6KYT
 
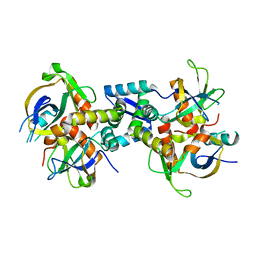 | | The structure of the M. tb toxin MazEF-mt1 complex | | Descriptor: | Antitoxin MazE9, Endoribonuclease MazF9 | | Authors: | Xie, W, Chen, R, Zhou, J. | | Deposit date: | 2019-09-20 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.00101161 Å) | | Cite: | Conserved Conformational Changes in the Regulation ofMycobacterium tuberculosisMazEF-mt1.
Acs Infect Dis., 6, 2020
|
|
7VA1
 
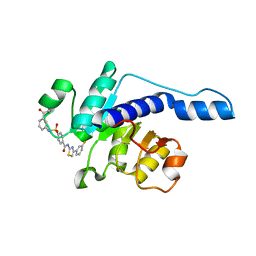 | |
8HKN
 
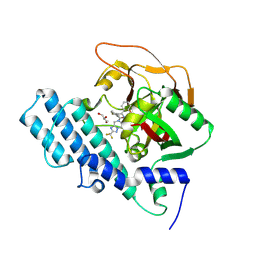 | |
8HLQ
 
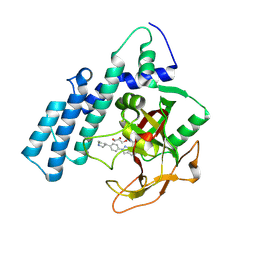 | |
8HLJ
 
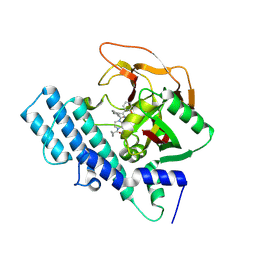 | |
8HKO
 
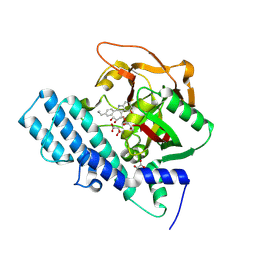 | |
8HKS
 
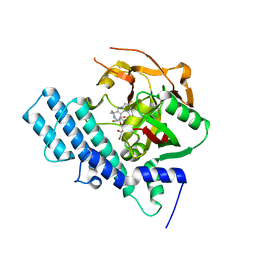 | | Mutated human ADP-ribosyltransferase 2 (PARP2) catalytic domain bound to Pamiparib(BGB-290) | | Descriptor: | (2R)-14-fluoro-2-methyl-6,9,10,19-tetrazapentacyclo[14.2.1.02,6.08,18.012,17]nonadeca-1(18),8,12(17),13,15-pentaen-11-one, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2022-11-28 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutated human ADP-ribosyltransferase 2 (PARP2) catalytic domain bound to Pamiparib
To Be Published
|
|
8HLR
 
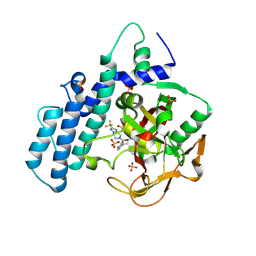 | |
7EJH
 
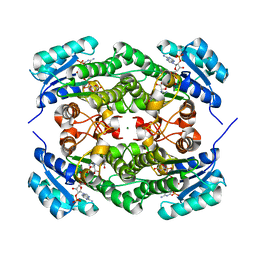 | | Crystal structure of KRED mutant-F147L/L153Q/Y190P/L199A/M205F/M206F and 2-hydroxyisoindoline-1,3-dione complex | | Descriptor: | 2-oxidanylisoindole-1,3-dione, 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase, MAGNESIUM ION, ... | | Authors: | Cui, J, Huang, X, Wang, B, Zhao, H, Zhou, J. | | Deposit date: | 2021-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72883928 Å) | | Cite: | Photoinduced chemomimetic biocatalysis for enantioselective intermolecular radical conjugate addition
Nat Catal, 2022
|
|
7EJJ
 
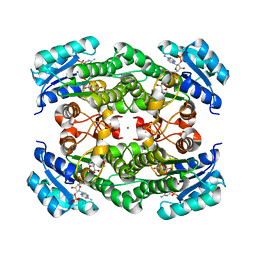 | | Crystal structure of KRED F147L/L153Q/Y190P variant and methyl methacrylate complex | | Descriptor: | 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cui, J, Huang, X, Wang, B, Zhao, H, Zhou, J. | | Deposit date: | 2021-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.80000663 Å) | | Cite: | Photoinduced chemomimetic biocatalysis for enantioselective intermolecular radical conjugate addition
Nat Catal, 2022
|
|
7EJI
 
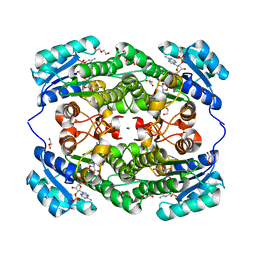 | | Crystal structure of KRED F147L/L153Q/Y190P/L199A/M205F/M206F variant and methyl methacrylate complex | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-oxidanylisoindole-1,3-dione, ... | | Authors: | Cui, J, Huang, X, Wang, B, Zhao, H, Zhou, J. | | Deposit date: | 2021-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.560016 Å) | | Cite: | Photoinduced chemomimetic biocatalysis for enantioselective intermolecular radical conjugate addition
Nat Catal, 2022
|
|
7CFM
 
 | | Cryo-EM structure of the P395-bound GPBAR-Gs complex | | Descriptor: | 2-(ethylamino)-6-[3-(4-propan-2-ylphenyl)propanoyl]-7,8-dihydro-5H-pyrido[4,3-d]pyrimidine-4-carboxamide, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | Authors: | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
7CFN
 
 | | Cryo-EM structure of the INT-777-bound GPBAR-Gs complex | | Descriptor: | (2S,4R)-4-[(3R,5S,6R,7R,8R,9S,10S,12S,13R,14S,17R)-6-ethyl-10,13-dimethyl-3,7,12-tris(oxidanyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-pentanoic acid, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | Authors: | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
7EA9
 
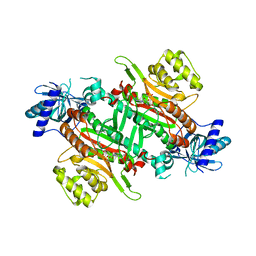 | | Crystal Structure of human lysyl-tRNA synthetase Y145H mutant | | Descriptor: | 5'-O-[(L-LYSYLAMINO)SULFONYL]ADENOSINE, GLYCEROL, Lysine--tRNA ligase | | Authors: | Wu, S, Hei, Z, Zheng, L, Zhou, J, Liu, Z, Wang, J, Fang, P. | | Deposit date: | 2021-03-06 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analyses of a human lysyl-tRNA synthetase mutant associated with autosomal recessive nonsyndromic hearing impairment.
Biochem.Biophys.Res.Commun., 554, 2021
|
|
7E2M
 
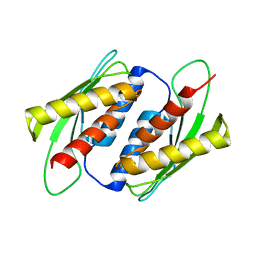 | | Crystal structure of the RWD domain of human GCN2 - 2 | | Descriptor: | eIF-2-alpha kinase GCN2 | | Authors: | Hei, Z, Zhou, J, Liu, Z, Wang, J, Fang, P. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures reveal a novel dimer of the RWD domain of human general control nonderepressible 2.
Biochem.Biophys.Res.Commun., 549, 2021
|
|
7E2K
 
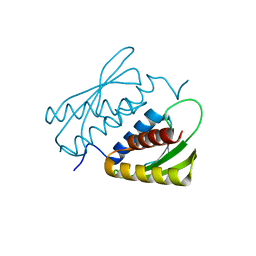 | | Crystal structure of the RWD domain of human GCN2 - 1 | | Descriptor: | eIF-2-alpha kinase GCN2 | | Authors: | Hei, Z, Zhou, J, Liu, Z, Wang, J, Fang, P. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.041 Å) | | Cite: | Crystal structures reveal a novel dimer of the RWD domain of human general control nonderepressible 2.
Biochem.Biophys.Res.Commun., 549, 2021
|
|
7FCC
 
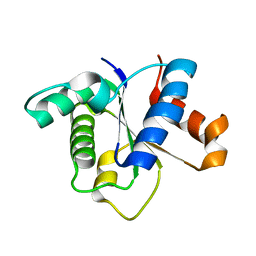 | | IL-1RAcPb TIR domain | | Descriptor: | Isoform 4 of Interleukin-1 receptor accessory protein | | Authors: | Wang, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Structural basis of the IL-1 receptor TIR domain-mediated IL-1 signaling
Iscience, 25, 2022
|
|
7FCJ
 
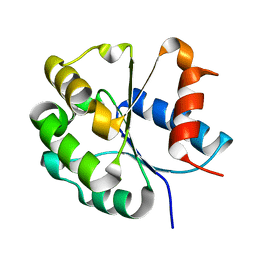 | | Zebrafish SIGIRR TIR domain mutant - C299S | | Descriptor: | SIGIRR protein | | Authors: | Wang, X, Zhou, J. | | Deposit date: | 2021-07-15 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural basis of the IL-1 receptor TIR domain-mediated IL-1 signaling
Iscience, 25, 2022
|
|
7FCH
 
 | | IL-18Rbeta TIR domain | | Descriptor: | Interleukin-18 receptor accessory protein | | Authors: | Wang, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | Structural basis of the IL-1 receptor TIR domain-mediated IL-1 signaling
Iscience, 25, 2022
|
|
7FD3
 
 | | IL-1RAPL2 TIR domain | | Descriptor: | X-linked interleukin-1 receptor accessory protein-like 2 | | Authors: | Wang, X, Zhou, J. | | Deposit date: | 2021-07-15 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis of the IL-1 receptor TIR domain-mediated IL-1 signaling
Iscience, 25, 2022
|
|
7FCL
 
 | | Zebrafish SIGIRR TIR domain | | Descriptor: | SIGIRR protein | | Authors: | Wang, X, Zhou, J. | | Deposit date: | 2021-07-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural basis of the IL-1 receptor TIR domain-mediated IL-1 signaling
Iscience, 25, 2022
|
|
