6ISL
 
 | | SnoaL-like cyclase XimE with its product xiamenmycin B | | Descriptor: | (2R,3S)-2-methyl-2-(4-methylpent-3-enyl)-3-oxidanyl-3,4-dihydrochromene-6-carboxylic acid, XimE, SnoaL-like domain protein | | Authors: | He, B, Zhou, T, Bu, X, Weng, J, Xu, J, Lin, S, Zheng, J, Zhao, Y, Xu, M. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Enzymatic Pyran Formation Involved in Xiamenmycin Biosynthesis
Acs Catalysis, 9, 2019
|
|
3NAO
 
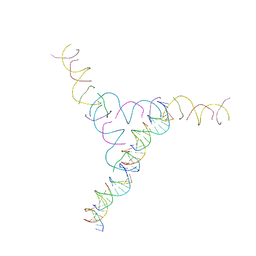 | | A DNA Crystal Designed to Contain Two Molecules per Asymmetric Unit Cell | | Descriptor: | DNA (5'-D(*AP*GP*CP*CP*TP*AP*CP*CP*TP*GP*CP*GP*TP*GP*GP*AP*CP*AP*GP*AP*C)-3'), DNA (5'-D(*CP*TP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), ... | | Authors: | Wang, T, Sha, R, Birktoft, J.J, Zheng, J, Mao, M, Seeman, N.C. | | Deposit date: | 2010-06-02 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (5.03 Å) | | Cite: | A DNA crystal designed to contain two molecules per asymmetric unit.
J.Am.Chem.Soc., 132, 2010
|
|
3GBI
 
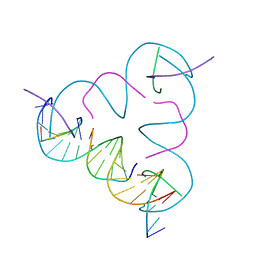 | | The Rational Design and Structural Analysis of a Self-Assembled Three-Dimensional DNA Crystal | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | Authors: | Birktoft, J.J, Zheng, J, Seeman, N.C. | | Deposit date: | 2009-02-19 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.018 Å) | | Cite: | From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal.
Nature, 461, 2009
|
|
2R28
 
 | |
4R2Y
 
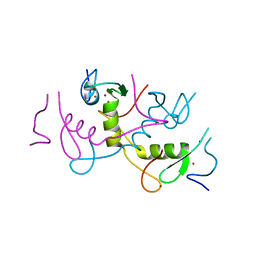 | | Crystal structure of APC11 RING domain | | Descriptor: | Anaphase-promoting complex subunit 11, ZINC ION | | Authors: | Brown, N.G, Watson, E.R, Weissmann, F, Jarvis, M.A, Vanderlinden, R, Grace, C.R.R, Frye, J.J, Dube, P, Qiao, R, Petzold, G, Cho, S.E, Alsharif, O, Bao, J, Zheng, J, Nourse, A, Kurinov, I, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Mechanism of Polyubiquitination by Human Anaphase-Promoting Complex: RING Repurposing for Ubiquitin Chain Assembly.
Mol.Cell, 56, 2014
|
|
8HVR
 
 | |
8IAA
 
 | |
8IA9
 
 | |
6KJ7
 
 | | E. coli ATCase catalytic subunit mutant - G166P | | Descriptor: | Aspartate carbamoyltransferase catalytic subunit | | Authors: | Lei, Z, Zheng, J, Jia, Z. | | Deposit date: | 2019-07-21 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.839 Å) | | Cite: | New regulatory mechanism-based inhibitors of aspartate transcarbamoylase for potential anticancer drug development.
Febs J., 287, 2020
|
|
6KJ9
 
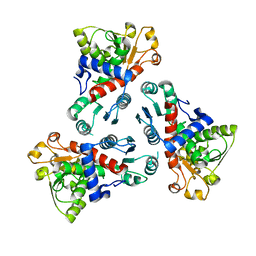 | | E. coli ATCase catalytic subunit mutant - G128/130A | | Descriptor: | Aspartate carbamoyltransferase catalytic subunit | | Authors: | Lei, Z, Zheng, J, Jia, Z.C. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New regulatory mechanism-based inhibitors of aspartate transcarbamoylase for potential anticancer drug development.
Febs J., 287, 2020
|
|
6KJ8
 
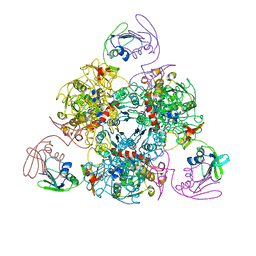 | | E. coli ATCase holoenzyme mutant - G166P (catalytic chain) | | Descriptor: | Aspartate carbamoyltransferase catalytic subunit, Aspartate carbamoyltransferase regulatory chain, ZINC ION | | Authors: | Lei, Z, Zheng, J, Jia, Z.C. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | New regulatory mechanism-based inhibitors of aspartate transcarbamoylase for potential anticancer drug development.
Febs J., 287, 2020
|
|
6KJA
 
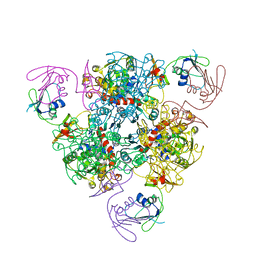 | | E. coli ATCase holoenzyme mutant - G128/130A (catalytic chain) | | Descriptor: | Aspartate carbamoyltransferase catalytic subunit, Aspartate carbamoyltransferase regulatory chain, ZINC ION | | Authors: | Lei, Z, Zheng, J, Jia, Z.C. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.064 Å) | | Cite: | New regulatory mechanism-based inhibitors of aspartate transcarbamoylase for potential anticancer drug development.
Febs J., 287, 2020
|
|
4P0E
 
 | |
5XWW
 
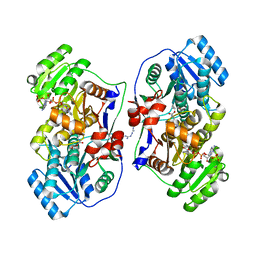 | |
5XWV
 
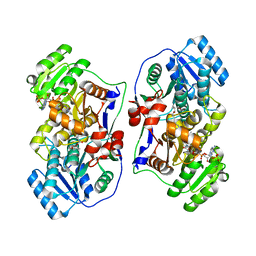 | |
2HRJ
 
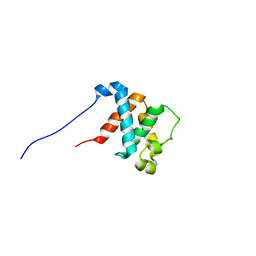 | |
3Q9G
 
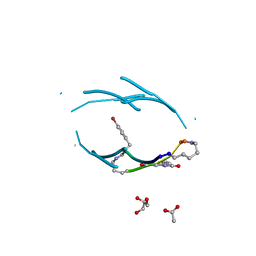 | | VQIVY segment from Alzheimer's tau displayed on 42-membered macrocycle scaffold | | Descriptor: | ACETIC ACID, Cyclic pseudo-peptide VQIV(4BF)(ORN)(HAO)KL(ORN), GLYCEROL | | Authors: | Liu, C, Sawaya, M.R, Eisenberg, D, Nowick, J.S, Cheng, P, Zheng, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Characteristics of Amyloid-Related Oligomers Revealed by Crystal Structures of Macrocyclic beta-Sheet Mimics.
J.Am.Chem.Soc., 133, 2011
|
|
3Q9J
 
 | | AIIFL segment derived from Alzheimer's Amyloid-Beta displayed on 42-membered macrocycle scaffold | | Descriptor: | CHLORIDE ION, Cyclic pseudo-peptide AIIFL(ORN)(HAO)YK(ORN), GLYCEROL, ... | | Authors: | Liu, C, Sawaya, M.R, Eisenberg, D, Nowick, J.S, Cheng, P, Zheng, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Characteristics of Amyloid-Related Oligomers Revealed by Crystal Structures of Macrocyclic beta-Sheet Mimics.
J.Am.Chem.Soc., 133, 2011
|
|
3Q9I
 
 | | LVFFA segment from Alzheimer's Amyloid-Beta displayed on 42-membered macrocycle scaffold, bromide derivative | | Descriptor: | CHLORIDE ION, Cyclic pseudo-peptide LV(4BF)FA(ORN)(HAO)LK(ORN), GLYCEROL, ... | | Authors: | Liu, C, Sawaya, M.R, Eisenberg, D, Nowick, J.S, Cheng, P, Zheng, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Characteristics of Amyloid-Related Oligomers Revealed by Crystal Structures of Macrocyclic beta-Sheet Mimics.
J.Am.Chem.Soc., 133, 2011
|
|
3Q9H
 
 | | LVFFA segment from Alzheimer's Amyloid-Beta displayed on 42-membered macrocycle scaffold | | Descriptor: | 1,4-BUTANEDIOL, Cyclic pseudo-peptide LVFFA(ORN)(HAO)LK(ORN), GLYCEROL, ... | | Authors: | Liu, C, Sawaya, M.R, Eisenberg, D, Nowick, J.S, Cheng, P, Zheng, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characteristics of Amyloid-Related Oligomers Revealed by Crystal Structures of Macrocyclic beta-Sheet Mimics.
J.Am.Chem.Soc., 133, 2011
|
|
2L6G
 
 | |
2L6H
 
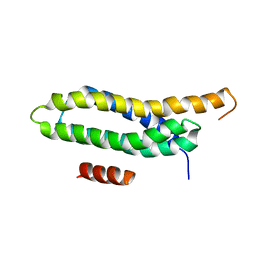 | |
2L6F
 
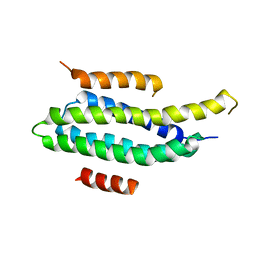 | | NMR Solution structure of FAT domain of FAK complexed with LD2 and LD4 motifs of PAXILLIN | | Descriptor: | Focal adhesion kinase 1, linker1, Paxillin, ... | | Authors: | Bertolucci, C.M, Guibao, C, Zhang, C, Zheng, J. | | Deposit date: | 2010-11-19 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Fat Domain of Fak Complexed with Ld2 and Ld4 Motifs of Paxillin
To be Published
|
|
6AD3
 
 | |
5EK7
 
 | |
