1HU8
 
 | | CRYSTAL STRUCTURE OF THE MOUSE P53 CORE DNA-BINDING DOMAIN AT 2.7A RESOLUTION | | Descriptor: | CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Zhao, K, Chai, X, Johnston, K, Clements, A, Marmorstein, R. | | Deposit date: | 2001-01-04 | | Release date: | 2001-07-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the mouse p53 core DNA-binding domain at 2.7 A resolution.
J.Biol.Chem., 276, 2001
|
|
7QK4
 
 | | EED in complex with PRC2 allosteric inhibitor compound 22 (MAK683) | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-[(5-fluoranyl-2,3-dihydro-1-benzofuran-4-yl)methyl]-8-(2-methylpyridin-3-yl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
7QJU
 
 | | EED in complex with PRC2 allosteric inhibitor compound 7 | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-[4-[(dimethylamino)methyl]phenyl]-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
7QJG
 
 | | EED in complex with PRC2 allosteric inhibitor compound 6 | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-(2,3-dihydro-1-benzofuran-7-ylmethyl)-8-[4-[(dimethylamino)methyl]phenyl]-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | Deposit date: | 2021-12-16 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
3TG5
 
 | | Structure of SMYD2 in complex with p53 and SAH | | Descriptor: | Cellular tumor antigen p53, GLYCEROL, N-lysine methyltransferase SMYD2, ... | | Authors: | Zhao, K, Wang, L. | | Deposit date: | 2011-08-17 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of human SMYD2 reveals the basis of p53 tumor suppressor methylation
J.Biol.Chem., 2011
|
|
1JIA
 
 | |
3TG4
 
 | | Structure of SMYD2 in complex with SAM | | Descriptor: | GLYCEROL, N-lysine methyltransferase SMYD2, S-ADENOSYLMETHIONINE, ... | | Authors: | Zhao, K, Wang, L. | | Deposit date: | 2011-08-17 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human SMYD2 reveals the basis of p53 tumor suppressor methylation
J.Biol.Chem., 2011
|
|
1SZD
 
 | | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD+-dependent Sir2 histone/protein deacetylases | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhao, K, Harshaw, R, Chai, X, Marmorstein, R. | | Deposit date: | 2004-04-05 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1SZC
 
 | | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD+-dependent Sir2 histone/protein deacetylases | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhao, K, Harshaw, R, Chai, X, Marmorstein, R. | | Deposit date: | 2004-04-05 | | Release date: | 2004-06-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1Q1A
 
 | | Structure of the yeast Hst2 protein deacetylase in ternary complex with 2'-O-acetyl ADP ribose and histone peptide | | Descriptor: | 2'-O-ACETYL ADENOSINE-5-DIPHOSPHORIBOSE, HST2 protein, Histone H4, ... | | Authors: | Zhao, K, Chai, X, Marmorstein, R. | | Deposit date: | 2003-07-18 | | Release date: | 2003-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the yeast Hst2 protein deacetylase in ternary complex with 2'-O-Acetyl ADP ribose and histone peptide.
Structure, 11, 2003
|
|
1Q17
 
 | | Structure of the yeast Hst2 protein deacetylase in ternary complex with 2'-O-acetyl ADP ribose and histone peptide | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHLORIDE ION, HST2 protein, ... | | Authors: | Zhao, K, Chai, X, Marmorstein, R. | | Deposit date: | 2003-07-18 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Yeast Hst2 Protein Deacetylase in Ternary Complex with 2'-O-Acetyl
ADP Ribose and Histone Peptide.
Structure, 11, 2003
|
|
6LO2
 
 | | Crystal structure of EED in complex with EZH2 peptide and compound 11# | | Descriptor: | 8-[4-[(dimethylamino)methyl]phenyl]-N-[(2-methoxyphenyl)methyl]-[1,2,4]triazolo[4,3-a]pyridin-5-amine, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Chen, Z. | | Deposit date: | 2020-01-02 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of EED in complex with EZH2 peptide and cmpd20
To Be Published
|
|
1NFJ
 
 | | Structure of a Sir2 substrate, alba, reveals a mechanism for deactylation-induced enhancement of DNA-binding | | Descriptor: | conserved hypothetical protein AF1956 | | Authors: | Zhao, K, Chai, X, Marmorstein, R. | | Deposit date: | 2002-12-15 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Sir2 substrate, alba, reveals a mechanism for deacetylation-induced enhancement of DNA-binding
J.Biol.Chem., 278, 2003
|
|
1NFH
 
 | | Structure of a Sir2 substrate, alba, reveals a mechanism for deactylation-induced enhancement of DNA-binding | | Descriptor: | conserved hypothetical protein AF1956 | | Authors: | Zhao, K, Chai, X, Marmorstein, R. | | Deposit date: | 2002-12-15 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of a Sir2 substrate, Alba, reveals a mechanism for deacetylation-induced enhancement of DNA-binding
J.Biol.Chem., 278, 2003
|
|
1Q14
 
 | | Structure and autoregulation of the yeast Hst2 homolog of Sir2 | | Descriptor: | CHLORIDE ION, HST2 protein, ZINC ION | | Authors: | Zhao, K, Chai, X, Clements, A, Marmorstein, R. | | Deposit date: | 2003-07-18 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and autoregulation of the Yeast Hst2 homolog of Sir2
Nat.Struct.Biol., 10, 2003
|
|
5H19
 
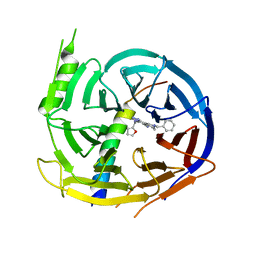 | | EED in complex with PRC2 allosteric inhibitor EED162 | | Descriptor: | 5-(furan-2-ylmethylamino)-9-(phenylmethyl)-8,10-dihydro-7H-[1,2,4]triazolo[3,4-a][2,7]naphthyridine-6-carbonitrile, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
1S5P
 
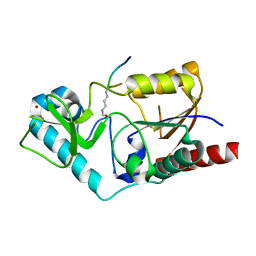 | | Structure and substrate binding properties of cobB, a Sir2 homolog protein deacetylase from Eschericia coli. | | Descriptor: | HISTONE H4 (RESIDUES 12-19), NAD-dependent deacetylase, ZINC ION | | Authors: | Zhao, K, Chai, X, Marmorstein, R. | | Deposit date: | 2004-01-21 | | Release date: | 2004-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure and Substrate Binding Properties of cobB, a Sir2 Homolog Protein Deacetylase from Eschericia coli.
J.Mol.Biol., 337, 2004
|
|
5H14
 
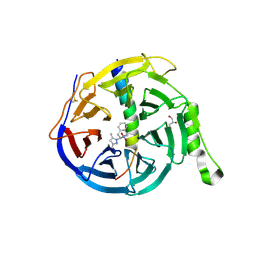 | | EED in complex with an allosteric PRC2 inhibitor EED666 | | Descriptor: | 2-[3-(3,5-dimethylpyrazol-1-yl)-4-nitro-phenyl]-3,4-dihydro-1H-isoquinoline, GLYCEROL, Histone-lysine N-methyltransferase EZH2, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
5H25
 
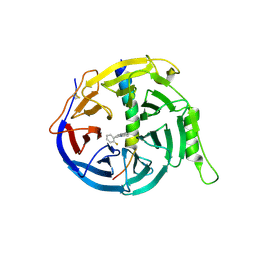 | | EED in complex with PRC2 allosteric inhibitor compound 11 | | Descriptor: | 5-(2-fluorophenyl)-2,3-dihydroimidazo[2,1-a]isoquinoline, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-14 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Discovery of First-in-Class, Potent, and Orally Bioavailable Embryonic Ectoderm Development (EED) Inhibitor with Robust Anticancer Efficacy
J. Med. Chem., 60, 2017
|
|
5H24
 
 | | EED in complex with PRC2 allosteric inhibitor compound 8 | | Descriptor: | 5-(furan-2-ylmethylamino)-[1,2,4]triazolo[4,3-a]pyridine-6-carbonitrile, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-14 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of First-in-Class, Potent, and Orally Bioavailable Embryonic Ectoderm Development (EED) Inhibitor with Robust Anticancer Efficacy
J. Med. Chem., 60, 2017
|
|
5GSA
 
 | | EED in complex with an allosteric PRC2 inhibitor | | Descriptor: | Histone-lysine N-methyltransferase EZH2, N-(furan-2-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of
Nat. Chem. Biol., 13, 2017
|
|
5H13
 
 | | EED in complex with PRC2 allosteric inhibitor EED396 | | Descriptor: | 4-azanylidene-2-(3-methoxy-4-propan-2-yloxy-phenyl)-6,7-dihydro-[1,3]benzodioxolo[6,5-a]quinolizine-3-carbonitrile, GLYCEROL, PRASEODYMIUM ION, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
5H17
 
 | | EED in complex with PRC2 allosteric inhibitor EED210 | | Descriptor: | (3R,4aS,10aS)-6-methoxy-3-[(3-methoxyphenyl)methyl]-1-methyl-3,4,4a,5,10,10a-hexahydro-2H-benzo[g]quinoline, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
5H15
 
 | | EED in complex with PRC2 allosteric inhibitor EED709 | | Descriptor: | (3R,4S)-1-[(2-methoxyphenyl)methyl]-N,N-dimethyl-4-(1-methylindol-3-yl)pyrrolidin-3-amine, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
8JH6
 
 | | Crystal structure of apixaban-bound racemic ABLE | | Descriptor: | 1-(4-METHOXYPHENYL)-7-OXO-6-[4-(2-OXOPIPERIDIN-1-YL)PHENYL]-4,5,6,7-TETRAHYDRO-1H-PYRAZOLO[3,4-C]PYRIDINE-3-CARBOXAMIDE, D-ABLE, L-ABLE | | Authors: | Zhao, K, Li, T. | | Deposit date: | 2023-05-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of apixaban-bound racemic ABLE
To Be Published
|
|
