7CMU
 
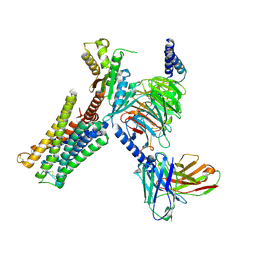 | | Dopamine Receptor D3R-Gi-Pramipexole complex | | Descriptor: | (6S)-N6-propyl-4,5,6,7-tetrahydro-1,3-benzothiazole-2,6-diamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Mao, C, Krumm, B, Zhou, X, Tan, Y, Huang, X.-P, Liu, Y, Shen, D.-D, Jiang, Y, Yu, X, Jiang, H, Melcher, K, Roth, B, Cheng, X, Zhang, Y, Xu, H. | | Deposit date: | 2020-07-29 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of the human dopamine D3 receptor-G i complexes.
Mol.Cell, 81, 2021
|
|
7CMV
 
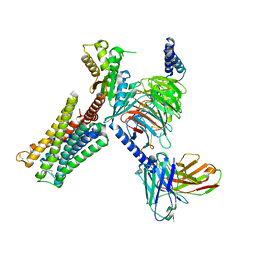 | | Dopamine Receptor D3R-Gi-PD128907 complex | | Descriptor: | (4aR,10bR)-4-propyl-3,4a,5,10b-tetrahydro-2H-chromeno[4,3-b][1,4]oxazin-9-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Mao, C, Krumm, B, Zhou, X, Tan, Y, Huang, X.-P, Liu, Y, Shen, D.-D, Jiang, Y, Yu, X, Jiang, H, Melcher, K, Roth, B, Cheng, X, Zhang, Y, Xu, H. | | Deposit date: | 2020-07-29 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of the human dopamine D3 receptor-G i complexes.
Mol.Cell, 81, 2021
|
|
7C72
 
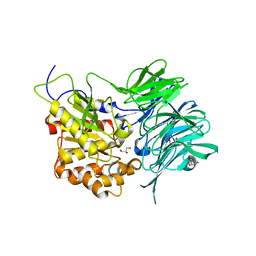 | | Structure of a mycobacterium tuberculosis puromycin-hydrolyzing peptidase | | Descriptor: | D-MALATE, GLYCEROL, Prolyl oligopeptidase | | Authors: | Ruiz-Carrillo, D, Zhao, Y.H, Feng, Q, Zhou, X, Zhang, Y, Jiang, J, Lukman, M. | | Deposit date: | 2020-05-22 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.00004458 Å) | | Cite: | Mycobacterium tuberculosis puromycin hydrolase displays a prolyl oligopeptidase fold and an acyl aminopeptidase activity.
Proteins, 89, 2021
|
|
7VMY
 
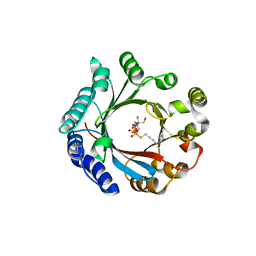 | | Crystal structure of LimF prenyltransferase bound with GSPP | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GERANYL S-THIOLODIPHOSPHATE, LynF/TruF/PatF family peptide O-prenyltransferase, ... | | Authors: | Hamada, K, Kobayashi, S, Okada, C, Zhang, Y, Inoue, S, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | LimF is a versatile prenyltransferase for histidine-C-geranylation on diverse non-natural substrates
Nat Catal, 2022
|
|
7VMW
 
 | | Crystal structure of LimF prenyltransferase bound with a peptide substrate and GSPP | | Descriptor: | GERANYL S-THIOLODIPHOSPHATE, LynF/TruF/PatF family peptide O-prenyltransferase, MAGNESIUM ION, ... | | Authors: | Hamada, K, Kobayashi, S, Okada, C, Zhang, Y, Inoue, S, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | LimF is a versatile prenyltransferase for histidine-C-geranylation on diverse non-natural substrates
Nat Catal, 2022
|
|
7Y1Y
 
 | | S-ECD (Omicron BA.2) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2022-06-09 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Basis for the Enhanced Infectivity and Immune Evasion of Omicron Subvariants.
Viruses, 15, 2023
|
|
7Y1Z
 
 | | S-ECD (Omicron BA.3) in complex with three PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2022-06-09 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis for the Enhanced Infectivity and Immune Evasion of Omicron Subvariants.
Viruses, 15, 2023
|
|
7Y76
 
 | | SIT1-ACE2-BA.5 RBD | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shen, Y.P, Li, Y.N, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2022-06-21 | | Release date: | 2023-01-04 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of ACE2-SIT1 recognized by Omicron variants of SARS-CoV-2.
Cell Discov, 8, 2022
|
|
7Y20
 
 | | S-ECD (Omicron BA.3) in complex with two PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2022-06-09 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Basis for the Enhanced Infectivity and Immune Evasion of Omicron Subvariants.
Viruses, 15, 2023
|
|
7Y21
 
 | | S-ECD (Omicron BA.5) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2022-06-09 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural Basis for the Enhanced Infectivity and Immune Evasion of Omicron Subvariants.
Viruses, 15, 2023
|
|
7VYU
 
 | |
7XNG
 
 | | Crystal structure of CBP bromodomain liganded with Y08092(31g) | | Descriptor: | 3-[(1-ethanoylindol-3-yl)carbonylamino]-5-[[(2S)-oxan-2-yl]oxymethyl]benzoic acid, CREB-binding protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Xiang, Q, Zhang, Y, Wang, C, Song, M, Xu, Y. | | Deposit date: | 2022-04-28 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of CBP bromodomain liganded with Y08092(31g)
To Be Published
|
|
7XZQ
 
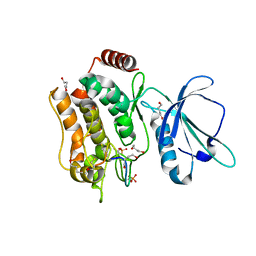 | | Crystal structure of TNIK-thiopeptide TP1 complex | | Descriptor: | 1,4-BUTANEDIOL, TRAF2 and NCK-interacting protein kinase, thiopeptide TP1 | | Authors: | Hamada, K, Vinogradov, A.A, Zhang, Y, Chang, J.S, Nishimura, H, Goto, Y, Onaka, H, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | De Novo Discovery of Thiopeptide Pseudo-natural Products Acting as Potent and Selective TNIK Kinase Inhibitors.
J.Am.Chem.Soc., 144, 2022
|
|
7XZR
 
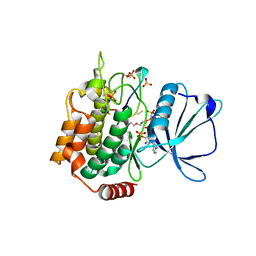 | | Crystal structure of TNIK-AMPPNP-thiopeptide TP15 complex | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION, ... | | Authors: | Hamada, K, Vinogradov, A.A, Zhang, Y, Chang, J.S, Nishimura, H, Goto, Y, Onaka, H, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | De Novo Discovery of Thiopeptide Pseudo-natural Products Acting as Potent and Selective TNIK Kinase Inhibitors.
J.Am.Chem.Soc., 144, 2022
|
|
7XUF
 
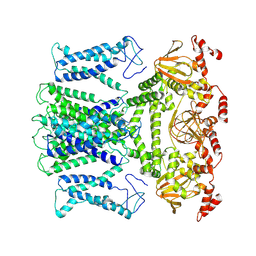 | | Cryo-EM structure of the AKT1-AtKC1 complex from Arabidopsis thaliana | | Descriptor: | POTASSIUM ION, Potassium channel AKT1, Potassium channel KAT3 | | Authors: | Yang, G.H, Lu, Y.M, Jia, Y.T, Yang, F, Zhang, Y.M, Xu, X, Li, X.M, Lei, J.L. | | Deposit date: | 2022-05-18 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
7Y75
 
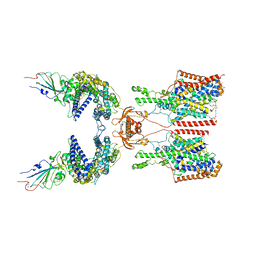 | | SIT1-ACE2-BA.2 RBD | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shen, Y.P, Li, Y.N, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2022-06-21 | | Release date: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of ACE2-SIT1 recognized by Omicron variants of SARS-CoV-2.
Cell Discov, 8, 2022
|
|
7CYQ
 
 | | Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Yan, L, Ge, J, Zheng, L, Zhang, Y, Gao, Y, Wang, T, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis.
Cell, 184, 2021
|
|
7E7L
 
 | | The crystal structure of arylacetate decarboxylase from Olsenella scatoligenes. | | Descriptor: | 4-HYDROXYPHENYLACETATE, Hydroxyphenylacetic acid decarboxylase | | Authors: | Lu, Q, Duan, Y, Zhang, Y, Yuchi, Z. | | Deposit date: | 2021-02-26 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | The Glycyl Radical Enzyme Arylacetate Decarboxylase from Olsenella scatoligenes
Acs Catalysis, 11, 2021
|
|
7WDM
 
 | | Fungal immunomodulatory protein FIP-gmi | | Descriptor: | Immunomodulatory protein | | Authors: | Liu, Y, Bastiaan-Net, S, Zhang, Y, Hoppenbrouwers, T, Xie, Y, Wang, Y, Wei, X, Du, G, Zhang, H, Imam, K.M.S.U, Wichers, H.J, Li, Z. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.123 Å) | | Cite: | Linking the thermostability of FIP-nha (Nectria haematococca) to its structural properties
To Be Published
|
|
7Y3Z
 
 | |
7VLZ
 
 | | Crystal structure of the collagenase unit of a Vibrio collagenase from Vibrio harveyi VHJR7 | | Descriptor: | CALCIUM ION, Peptide P1, Peptide P2, ... | | Authors: | Cao, H.Y, Wang, Y, Peng, M, Zhang, Y.Z. | | Deposit date: | 2021-10-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the collagenase unit of a Vibrio collagenase from Vibrio harveyi VHJR7
To Be Published
|
|
