7EO8
 
 | | Crystal structure of SARS coronavirus main protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2808516 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
2O3J
 
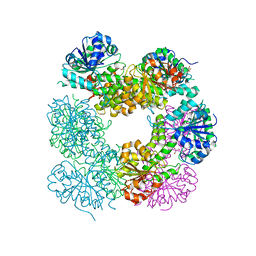 | | Structure of Caenorhabditis Elegans UDP-Glucose Dehydrogenase | | Descriptor: | GLYCEROL, UDP-glucose 6-dehydrogenase | | Authors: | Zhang, Y, Zhan, C, Patskovsky, Y, Ramagopal, U, Shi, W, Toro, R, Wengerter, B.C, Milst, S, Vidal, M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-12-01 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of Caenorhabditis Elegans Udp-Glucose Dehydrogenase
To be Published
|
|
7DQZ
 
 | | Crystal structure of SARS 3C-like protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-24 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
6KQD
 
 | | Thermus thermophilus initial transcription complex comprising sigma A and 5'-OH RNA of 3 nt | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*CP*AP*GP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G*)-3'), ... | | Authors: | Zhang, Y, Li, L, Ebright, R.H. | | Deposit date: | 2019-08-17 | | Release date: | 2020-03-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3V7T
 
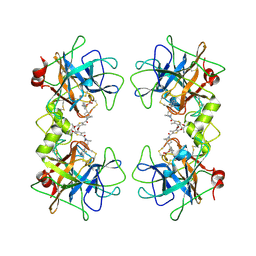 | |
4OIN
 
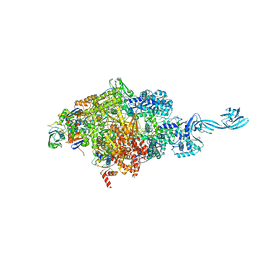 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077 | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
5YGF
 
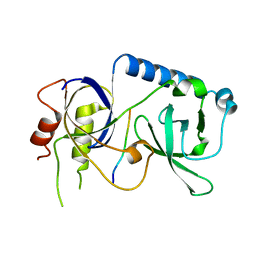 | |
5YGD
 
 | |
5YGB
 
 | |
5YGC
 
 | |
8HIC
 
 | | Crystal structure of UrtA from Prochlorococcus marinus str. MIT 9313 in complex with urea and calcium | | Descriptor: | CALCIUM ION, Putative urea ABC transporter, substrate binding protein, ... | | Authors: | Zhang, Y.Z, Wang, P, Wang, C. | | Deposit date: | 2022-11-19 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and molecular basis for urea recognition by Prochlorococcus.
J.Biol.Chem., 299, 2023
|
|
3TG3
 
 | | Crystal structure of the MAPK binding domain of MKP7 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein phosphatase 16 | | Authors: | Zhang, Y.Y, Liu, X, Wu, J.W, Wang, Z.X. | | Deposit date: | 2011-08-17 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.675 Å) | | Cite: | A Distinct Interaction Mode Revealed by the Crystal Structure of the Kinase p38alpha with the MAPK Binding Domain of the Phosphatase MKP5.
Sci.Signal., 4, 2011
|
|
3TG1
 
 | |
7VVW
 
 | | MmtN-SAM complex | | Descriptor: | GLYCEROL, PHOSPHATE ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Zhang, Y.Z, Peng, M, Li, C.Y. | | Deposit date: | 2021-11-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Insights into methionine S-methylation in diverse organisms.
Nat Commun, 13, 2022
|
|
7VVX
 
 | | MmtN-SAH-Met complex | | Descriptor: | METHIONINE, PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zhang, Y.Z, Peng, M, Li, C.Y. | | Deposit date: | 2021-11-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Insights into methionine S-methylation in diverse organisms.
Nat Commun, 13, 2022
|
|
5X21
 
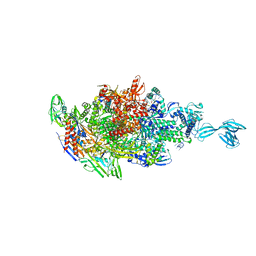 | | Crystal structure of Thermus thermophilus transcription initiation complex with GpA and pseudouridimycin (PUM) | | Descriptor: | (1S)-1,4-anhydro-5-[(N-carbamimidoylglycyl-N~2~-hydroxy-L-glutaminyl)amino]-5-deoxy-1-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)-D-ribitol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Zhang, Y, Ebright, R. | | Deposit date: | 2017-01-29 | | Release date: | 2017-07-05 | | Last modified: | 2022-10-12 | | Method: | X-RAY DIFFRACTION (3.323 Å) | | Cite: | Antibacterial Nucleoside-Analog Inhibitor of Bacterial RNA Polymerase.
Cell, 169, 2017
|
|
5X22
 
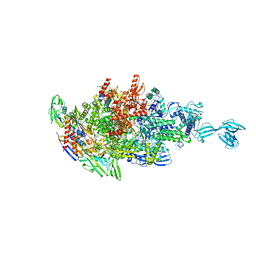 | |
7EU7
 
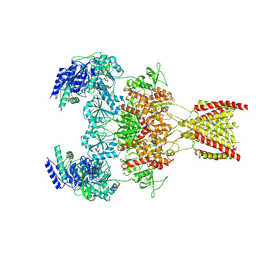 | | Structure of the human GluN1-GluN2A NMDA receptor in complex with S-ketamine, glycine and glutamate | | Descriptor: | (2~{S})-2-(2-chlorophenyl)-2-(methylamino)cyclohexan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, Y, Zhang, T, Zhu, S. | | Deposit date: | 2021-05-16 | | Release date: | 2021-08-04 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of ketamine action on human NMDA receptors.
Nature, 596, 2021
|
|
6A37
 
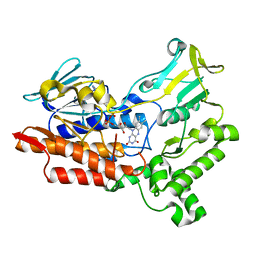 | |
8XRY
 
 | |
8XNG
 
 | |
8XS4
 
 | |
8XS5
 
 | |
8XAJ
 
 | | Cryo-EM structure of OSCA1.2-liposome-inside-in open state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Calcium permeable stress-gated cation channel 1 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XS0
 
 | |
