1G5J
 
 | | COMPLEX OF BCL-XL WITH PEPTIDE FROM BAD | | Descriptor: | APOPTOSIS REGULATOR BCL-X, BAD PROTEIN | | Authors: | Petros, A.M, Nettesheim, D.G, Wang, Y, Olejniczak, E.T, Meadows, R.P, Mack, J, Swift, K, Matayoshi, E.D, Zhang, H, Thompson, C.B, Fesik, S.W. | | Deposit date: | 2000-11-01 | | Release date: | 2001-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Rationale for Bcl-xL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies.
Protein Sci., 9, 2000
|
|
6DCW
 
 | |
5Z2T
 
 | | Crystal structure of DNA-bound DUX4-HD2 | | Descriptor: | 5'-D(*TP*TP*CP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*TP*T)-3', 5'-D(P*AP*AP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*GP*T)-3', Double homeobox protein 4 | | Authors: | Dong, X, Zhang, W, Wu, H, Huang, J, Zhang, M, Wang, P, Zhang, H, Chen, Z, Chen, S, Meng, G. | | Deposit date: | 2018-01-04 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.623 Å) | | Cite: | Structural basis of DUX4/IGH-driven transactivation.
Leukemia, 32, 2018
|
|
1M32
 
 | | Crystal Structure of 2-aminoethylphosphonate Transaminase | | Descriptor: | 2-aminoethylphosphonate-pyruvate aminotransferase, PHOSPHATE ION, PHOSPHONOACETALDEHYDE, ... | | Authors: | Chen, C.C.H, Zhang, H, Kim, A.D, Howard, A, Sheldrick, G.M, Mariano-Dunnaway, D, Herzberg, O. | | Deposit date: | 2002-06-26 | | Release date: | 2002-11-20 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Degradation Pathway of the Phosphonate Ciliatine: Crystal Structure of 2-Aminoethylphosphonate Transaminase
Biochemistry, 41, 2002
|
|
7F70
 
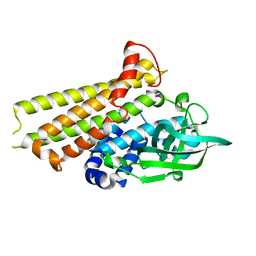 | | Crystal structure of Rv3094c | | Descriptor: | Rv3094c | | Authors: | Wang, Z.X, Ouyang, S.Y, Zhang, H.T. | | Deposit date: | 2021-06-26 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Omics analysis of Mycobacterium tuberculosis isolates uncovers Rv3094c, an ethionamide metabolism-associated gene.
Commun Biol, 6, 2023
|
|
5E6N
 
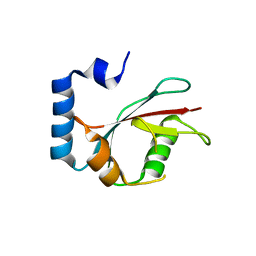 | | Crystal structure of C. elegans LGG-2 | | Descriptor: | Protein lgg-2 | | Authors: | Qi, X, Ren, J.Q, Wu, F, Zhang, H, Feng, W. | | Deposit date: | 2015-10-10 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy
Mol.Cell, 60, 2015
|
|
6V4L
 
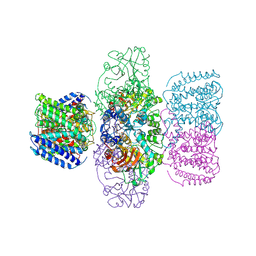 | | Structure of TrkH-TrkA in complex with ATPgammaS | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Potassium uptake protein TrkA, Trk system potassium uptake protein TrkH | | Authors: | Zhou, M, Zhang, H. | | Deposit date: | 2019-11-27 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | TrkA undergoes a tetramer-to-dimer conversion to open TrkH which enables changes in membrane potential.
Nat Commun, 11, 2020
|
|
5JPU
 
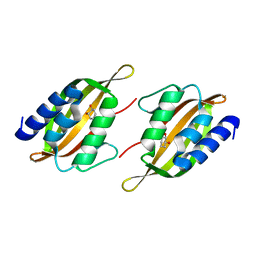 | | Structure of limonene epoxide hydrolase mutant - H-2-H5 complex with (S,S)-cyclohexane-1,2-diol | | Descriptor: | (1S,2S)-cyclohexane-1,2-diol, limonene epoxide hydrolase | | Authors: | Li, G, Zhang, H, Sun, Z, Liu, X, Reetz, M.T. | | Deposit date: | 2016-05-04 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Multi-Parameter Optimization in Directed Evolution: Engineering Thermostability, Enantioselectivity and Activity of an Epoxide Hydrolase
To be published
|
|
1W5X
 
 | | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | | Descriptor: | (2R,3R,4R,5R)-2,5-BIS[(2,3-DIFLUOROBENZYL)OXY]-3,4-DIHYDROXY-N,N'-BIS[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]HEXAN EDIAMIDE, POL POLYPROTEIN | | Authors: | Lindberg, J, Pyring, D, Loewgren, S, Rosenquist, A, Zuccarello, G, Kvarnstroem, I, Zhang, H, Vrang, L, Claesson, B, Hallberg, A, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-10 | | Release date: | 2004-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Symmetric Fluoro-Substituted Diol-Based HIV Protease Inhibitors. Ortho-Fluorinated and Meta-Fluorinated P1/P1'-Benzyloxy Side Groups Significantly Improve the Antiviral Activity and Preserve Binding Efficacy
Eur.J.Biochem., 271, 2004
|
|
5JPP
 
 | | Structure of limonene epoxide hydrolase mutant - H-2-H5 | | Descriptor: | limonene epoxide hydrolase | | Authors: | Li, G, Zhang, H, Sun, Z, Liu, X, Reetz, M.T. | | Deposit date: | 2016-05-04 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multi-Parameter Optimization in Directed Evolution: Engineering Thermostability, Enantioselectivity and Activity of an Epoxide Hydrolase
To be published
|
|
5F21
 
 | | human CD38 in complex with nanobody MU375 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Hao, Q, Zhang, H. | | Deposit date: | 2015-12-01 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Immuno-targeting the multifunctional CD38 using nanobody
Sci Rep, 6, 2016
|
|
8GQ5
 
 | |
1W5W
 
 | | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | | Descriptor: | (2R,3R,4R,5R)-2,5-BIS[(2,4-DIFLUOROBENZYL)OXY]-3,4-DIHYDROXY-N,N'-BIS[(1R,2S)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]HEXAN EDIAMIDE, POL POLYPROTEIN | | Authors: | Lindberg, J, Pyring, D, Loewgren, S, Rosenquist, A, Zuccarello, G, Kvarnstroem, I, Zhang, H, Vrang, L, Claesson, B, Hallberg, A, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-10 | | Release date: | 2004-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Symmetric Fluoro-Substituted Diol-Based HIV Protease Inhibitors. Ortho-Fluorinated and Meta-Fluorinated P1/P1'-Benzyloxy Side Groups Significantly Improve the Antiviral Activity and Preserve Binding Efficacy
Eur.J.Biochem., 271, 2004
|
|
1W5Y
 
 | | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | | Descriptor: | (2R,3R,4R,5R)-2,5-BIS[(2,5-DIFLUOROBENZYL)OXY]-3,4-DIHYDROXY-N,N'-BIS[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]HEXANEDIAMIDE, POL POLYPROTEIN | | Authors: | Lindberg, J, Pyring, D, Loewgren, S, Rosenquist, A, Zuccarello, G, Kvarnstroem, I, Zhang, H, Vrang, L, Claesson, B, Hallberg, A, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-10 | | Release date: | 2004-10-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Symmetric Fluoro-Substituted Diol-Based HIV Protease Inhibitors. Ortho-Fluorinated and Meta-Fluorinated P1/P1'-Benzyloxy Side Groups Significantly Improve the Antiviral Activity and Preserve Binding Efficacy
Eur.J.Biochem., 271, 2004
|
|
1RQX
 
 | | Crystal structure of ACC Deaminase complexed with Inhibitor | | Descriptor: | 1-AMINOCYCLOPROPYLPHOSPHONATE, 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Karthikeyan, S, Zhao, Z, Kao, C.L, Zhou, Q, Tao, Z, Zhang, H, Liu, H.W. | | Deposit date: | 2003-12-07 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of 1-aminocyclopropane-1-carboxylate deaminase: observation of an aminyl intermediate and identification of Tyr 294 as the active-site nucleophile.
Angew.Chem.Int.Ed.Engl., 43, 2004
|
|
1XA7
 
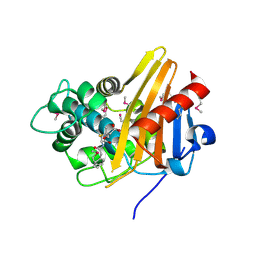 | | Crystal structure of the benzylpenicillin-acylated BlaR1 sensor domain from Staphylococcus aureus | | Descriptor: | OPEN FORM - PENICILLIN G, Regulatory protein BlaR1 | | Authors: | Wilke, M.S, Hills, T.L, Zhang, H.Z, Chambers, H.F, Strynadka, N.C. | | Deposit date: | 2004-08-25 | | Release date: | 2004-09-21 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the Apo and penicillin-acylated forms of the BlaR1 beta-lactam sensor of Staphylococcus aureus.
J.Biol.Chem., 279, 2004
|
|
3WOH
 
 | |
1YSW
 
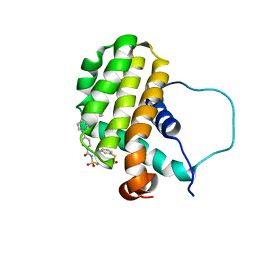 | | Solution structure of the anti-apoptotic protein Bcl-2 complexed with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-2 | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1YSI
 
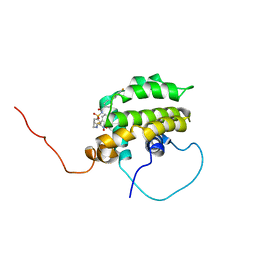 | | Solution structure of the anti-apoptotic protein Bcl-xL in complex with an acyl-sulfonamide-based ligand | | Descriptor: | Apoptosis regulator Bcl-X, N-[(4'-FLUORO-1,1'-BIPHENYL-4-YL)CARBONYL]-3-NITRO-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-08 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1YSG
 
 | | Solution Structure of the Anti-apoptotic Protein Bcl-xL in Complex with "SAR by NMR" Ligands | | Descriptor: | 4'-FLUORO-1,1'-BIPHENYL-4-CARBOXYLIC ACID, 5,6,7,8-TETRAHYDRONAPHTHALEN-1-OL, Apoptosis regulator Bcl-X | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-08 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1YSX
 
 | | Solution structure of domain 3 from human serum albumin complexed to an anti-apoptotic ligand directed against Bcl-xL and Bcl-2 | | Descriptor: | 4-({2-[(2,4-DIMETHYLPHENYL)SULFANYL]ETHYL}AMINO)-N-[(4'-FLUORO-1,1'-BIPHENYL-4-YL)CARBONYL]-3-NITROBENZENESULFONAMIDE, Serum albumin | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1YSN
 
 | | Solution structure of the anti-apoptotic protein Bcl-xL complexed with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-X | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-08 | | Release date: | 2005-06-07 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
7Y35
 
 | | Cryo-EM structure of the Abaloparatide-bound human PTH1R-Gs complex | | Descriptor: | Abaloparatide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhai, X, Mao, C, Shen, Q, Zang, S, Shen, D, Zhang, H, Chen, Z, Wang, G, Zhang, C, Zhang, Y, Liu, Z. | | Deposit date: | 2022-06-09 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of the Abaloparatide-bound human PTH1R-Gs complex
To Be Published
|
|
1IK1
 
 | | Solution Structure of an RNA Hairpin from HRV-14 | | Descriptor: | 5'-R(*GP*GP*UP*AP*CP*UP*AP*UP*GP*UP*AP*CP*CP*A)-3' | | Authors: | Huang, H, Alexandrov, A, Chen, X, Barnes III, T.W, Zhang, H, Dutta, K, Pascal, S.M. | | Deposit date: | 2001-05-01 | | Release date: | 2001-07-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA hairpin from HRV-14.
Biochemistry, 40, 2001
|
|
8GNI
 
 | | Human SARM1 bounded with NMN and Nanobody-C6, Conformation 1 | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1, Nanobody C6 | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | A conformation-specific nanobody targeting the nicotinamide mononucleotide-activated state of SARM1.
Nat Commun, 13, 2022
|
|
