6Y9V
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-8,13) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3H4P
 
 | | Proteasome 20S core particle from Methanocaldococcus jannaschii | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Jeffrey, P.D, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-20 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
8Y6O
 
 | | Cryo-EM Structure of the human minor pre-B complex (pre-precatalytic spliceosome) U11 and tri-snRNP part | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Centrosomal AT-AC splicing factor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Bai, R, Yuan, M, Zhang, P, Luo, T, Shi, Y, Wan, R. | | Deposit date: | 2024-02-02 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis of U12-type intron engagement by the fully assembled human minor spliceosome.
Science, 383, 2024
|
|
6Y9Y
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-7,13) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Y9W
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,8) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5Z6P
 
 | | The crystal structure of an agarase, AgWH50C | | Descriptor: | B-agarase | | Authors: | Mao, X, Zhou, J, Zhang, P, Zhang, L, Zhang, J, Li, Y. | | Deposit date: | 2018-01-24 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.061 Å) | | Cite: | Structure-based design of agarase AgWH50C from Agarivorans gilvus WH0801 to enhance thermostability.
Appl. Microbiol. Biotechnol., 103, 2019
|
|
7CX4
 
 | | Cryo-EM structure of the Evatanepag-bound EP2-Gs complex | | Descriptor: | 2-[3-[[(4-~{tert}-butylphenyl)methyl-pyridin-3-ylsulfonyl-amino]methyl]phenoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
7CX3
 
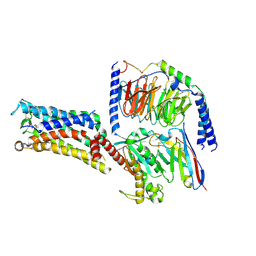 | | Cryo-EM structure of the Taprenepag-bound EP2-Gs complex | | Descriptor: | 2-[3-[[(4-pyrazol-1-ylphenyl)methyl-pyridin-3-ylsulfonyl-amino]methyl]phenoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
7CX2
 
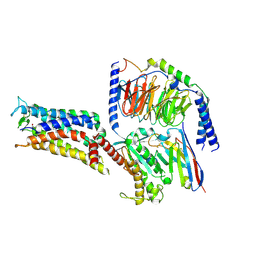 | | Cryo-EM structure of the PGE2-bound EP2-Gs complex | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
5X41
 
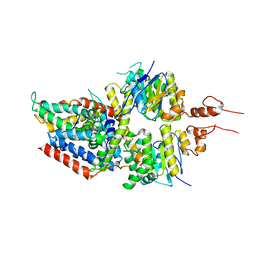 | | 3.5A resolution structure of a cobalt energy-coupling factor transporter using LCP method-CbiMQO | | Descriptor: | Cobalt ABC transporter ATP-binding protein, Cobalt transport protein CbiM, Uncharacterized protein CbiQ | | Authors: | Bao, Z, Qi, X, Zhao, W, Li, D, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
5X40
 
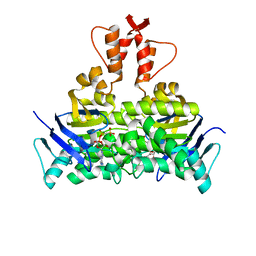 | | Structure of a CbiO dimer bound with AMPPCP | | Descriptor: | Cobalt ABC transporter ATP-binding protein, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Bao, Z, Qi, X, Wang, J, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
5XN6
 
 | | Heterodimer crystal structure of geranylgeranyl diphosphate synthases 1 with GGPPS Recruiting Protein(OsGRP) from Oryza sativa | | Descriptor: | Os02g0668100 protein, Os07g0580900 protein | | Authors: | Wang, C, Zhou, F, Lu, S, Zhang, P. | | Deposit date: | 2017-05-18 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.598 Å) | | Cite: | A recruiting protein of geranylgeranyl diphosphate synthase controls metabolic flux toward chlorophyll biosynthesis in rice
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5X3X
 
 | | 2.8A resolution structure of a cobalt energy-coupling factor transporter-CbiMQO | | Descriptor: | Cobalt ABC transporter ATP-binding protein, Cobalt transport protein CbiM, Uncharacterized protein CbiQ | | Authors: | Bao, Z, Qi, X, Wang, J, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.788 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
5XN5
 
 | |
4Z7F
 
 | | Crystal structure of FolT bound with folic acid | | Descriptor: | FOLIC ACID, Folate ECF transporter | | Authors: | Zhao, Q, Wang, C.C, Wang, C.Y, Zhang, P. | | Deposit date: | 2015-04-07 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Structures of FolT in substrate-bound and substrate-released conformations reveal a gating mechanism for ECF transporters
Nat Commun, 6, 2015
|
|
6ZDJ
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,10) | | Descriptor: | Gag protein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-06-14 | | Release date: | 2020-08-19 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZET
 
 | | Crystal structure of proteinase K nanocrystals by electron diffraction with a 20 micrometre C2 condenser aperture | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Evans, G, Zhang, P, Beale, E.V, Waterman, D.G. | | Deposit date: | 2020-06-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.701 Å) | | Cite: | A Workflow for Protein Structure Determination From Thin Crystal Lamella by Micro-Electron Diffraction.
Front Mol Biosci, 7, 2020
|
|
6ZEV
 
 | | Crystal structure of proteinase K lamellae by electron diffraction with a 20 micrometre C2 condenser aperture | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Evans, G, Zhang, P, Beale, E.V, Waterman, D.G. | | Deposit date: | 2020-06-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.4 Å) | | Cite: | A Workflow for Protein Structure Determination From Thin Crystal Lamella by Micro-Electron Diffraction.
Front Mol Biosci, 7, 2020
|
|
6ZEU
 
 | | Crystal structure of proteinase K lamella by electron diffraction with a 50 micrometre C2 condenser aperture | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Evans, G, Zhang, P, Beale, E.V, Waterman, D.G. | | Deposit date: | 2020-06-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.004 Å) | | Cite: | A Workflow for Protein Structure Determination From Thin Crystal Lamella by Micro-Electron Diffraction.
Front Mol Biosci, 7, 2020
|
|
5UP4
 
 | |
8FEC
 
 | | Structure of J-PKAc chimera complexed with Aplithianine derivative | | Descriptor: | 6-[(6P)-6-(4-bromo-1-methyl-1H-imidazol-5-yl)-2,3-dihydro-4H-1,4-thiazin-4-yl]-7H-purine, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Du, L, Wilson, B.A.P, Li, N, Martinez Fiesco, J.A, Dalilian, M, Wang, D, Smith, E.A, Wamiru, A, Goncharova, E.I, Zhang, P, O'Keefe, B.R. | | Deposit date: | 2022-12-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and Synthesis of a Naturally Derived Protein Kinase Inhibitor that Selectively Inhibits Distinct Classes of Serine/Threonine Kinases.
J.Nat.Prod., 86, 2023
|
|
8FE2
 
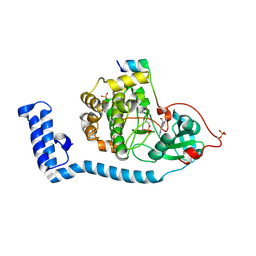 | | Structure of J-PKAc chimera complexed with Aplithianine A | | Descriptor: | 6-[(6M)-6-(1-methyl-1H-imidazol-5-yl)-2,3-dihydro-4H-1,4-thiazin-4-yl]-9H-purine, DnaJ homolog subfamily B member 1, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Du, L, Wilson, B.A.P, Li, N, Dalilian, M, Wang, D, Martinez Fiesco, J.A, Smith, E.A, Wamiru, A, Goncharova, E.I, Zhang, P, O'Keefe, B.R. | | Deposit date: | 2022-12-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Discovery and Synthesis of a Naturally Derived Protein Kinase Inhibitor that Selectively Inhibits Distinct Classes of Serine/Threonine Kinases.
J.Nat.Prod., 86, 2023
|
|
3J3Q
 
 | |
3JA6
 
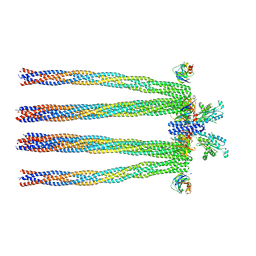 | | Cryo-electron Tomography and All-atom Molecular Dynamics Simulations Reveal a Novel Kinase Conformational Switch in Bacterial Chemotaxis Signaling | | Descriptor: | Chemotaxis protein CheA, Chemotaxis protein CheW, Methyl-accepting chemotaxis protein 2 | | Authors: | Cassidy, C.K, Himes, B.A, Alvarez, F.J, Ma, J, Zhao, G, Perilla, J.R, Schulten, K, Zhang, P. | | Deposit date: | 2015-04-21 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12.7 Å) | | Cite: | CryoEM and computer simulations reveal a novel kinase conformational switch in bacterial chemotaxis signaling.
Elife, 4, 2015
|
|
2M6I
 
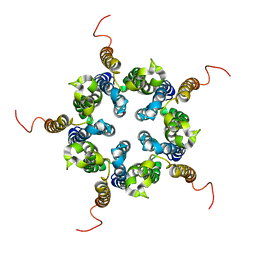 | | Putative pentameric open-channel structure of full-length transmembrane domains of human glycine receptor alpha1 subunit | | Descriptor: | Full-Length Transmembrane Domains of Human Glycine Receptor alpha1 Subunit | | Authors: | Mowrey, D, Cui, T, Jia, Y, Ma, D, Makhov, A.M, Zhang, P, Tang, P, Xu, Y. | | Deposit date: | 2013-03-29 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Open-Channel Structures of the Human Glycine Receptor alpha 1 Full-Length Transmembrane Domain.
Structure, 21, 2013
|
|
