4DVA
 
 | | The crystal structure of human urokinase-type plasminogen activator catalytic domain | | Descriptor: | HEXAETHYLENE GLYCOL, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Jiang, L, Botkjaer, K.A, Andersen, L.M, Yuan, C, Andreasen, P.A, Huang, M. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Rezymogenation of active urokinase induced by an inhibitory antibody
Biochem.J., 449, 2013
|
|
3KHV
 
 | | Crystal Structures of Urokinase-type Plasminogen Activator in Complex with 4-(Aminomethyl) Benzoic Acid and 4-(Aminomethyl-phenyl)-methanol | | Descriptor: | SULFATE ION, TRIETHYLENE GLYCOL, Urokinase-type plasminogen activator, ... | | Authors: | Jiang, L.-G, Zhao, G.-X, Bian, X.-B, Yuan, C, Huang, Z.-X, Huang, M.-D. | | Deposit date: | 2009-10-30 | | Release date: | 2009-12-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structures of Urokinase-type Plasminogen Activator in Complex with 4-(Aminomethyl) Benzoic Acid and 4-(Aminomethyl-phenyl)-methanol
CHIN.J.STRUCT.CHEM., 28, 2009
|
|
3KID
 
 | | The Crystal Structures of 2-Aminobenzothiazole-based Inhibitors in Complexes with Urokinase-type Plasminogen Activator | | Descriptor: | Urokinase-type plasminogen activator, ethyl 2-amino-1,3-benzothiazole-6-carboxylate | | Authors: | Jiang, L.-G, Yu, H.Y, Yuan, C, Wang, J.D, Chen, L.Q, Meehan, E.J, Huang, Z.-X, Huang, M.-D. | | Deposit date: | 2009-11-01 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal Structures of 2-Aminobenzothiazole-based Inhibitors in Complexes with Urokinase-type Plasminogen Activator
CHIN.J.STRUCT.CHEM., 28, 2009
|
|
3KGP
 
 | | Crystal Structures of Urokinase-type Plasminogen Activator in Complex with 4-(Aminomethyl) Benzoic Acid and 4-(Aminomethyl-phenyl)-methanol | | Descriptor: | 4-(aminomethyl)benzoic acid, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Jiang, L.-G, Zhao, G.-X, Bian, X.-B, Yuan, C, Huang, Z.-X, Huang, M.-D. | | Deposit date: | 2009-10-29 | | Release date: | 2009-12-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structures of Urokinase-type Plasminogen Activator in Complex with 4-(Aminomethyl) Benzoic Acid and 4-(Aminomethyl-phenyl)-methanol
CHIN.J.STRUCT.CHEM., 28, 2009
|
|
4XSK
 
 | | Structure of PAItrap, an uPA mutant | | Descriptor: | GLYCEROL, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Gong, L, Proulle, V, Hong, Z, Lin, Z, Liu, M, Yuan, C, Lin, L, Furie, B, Flaumenhaft, R, Andreasen, P, Furie, B, Huang, M. | | Deposit date: | 2015-01-22 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of PAItrap, an uPA mutant
To Be Published
|
|
8Y7F
 
 | |
8Y6Z
 
 | |
8Y7G
 
 | | Crystal structure of the Marinitoga sp. Csx1-Crn2 H495A mutant in complex with cyclic-tetraadenylate (cA4) | | Descriptor: | ACETATE ION, CRISPR-associated protein, RNA (5'-R(P*AP*A)-3'), ... | | Authors: | Zhang, D, Yuan, C, Lin, Z. | | Deposit date: | 2024-02-04 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural insight into the Csx1-Crn2 fusion self-limiting ribonuclease of type III CRISPR system.
Nucleic Acids Res., 52, 2024
|
|
6XVD
 
 | | Crystal structure of complex of urokinase and a upain-1 variant(W3F) in pH7.4 condition | | Descriptor: | Urokinase-type plasminogen activator, upain-1-W3F | | Authors: | Xue, G.P, Xie, X, Zhou, Y, Yuan, C, Huang, M.D, Jiang, L.G. | | Deposit date: | 2020-01-21 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insight to the residue in P2 position prevents the peptide inhibitor from being hydrolyzed by serine proteases.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
8Y75
 
 | |
4QTH
 
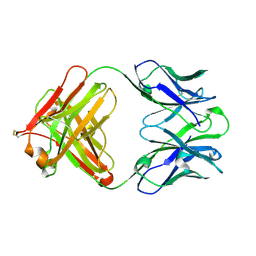 | | Crystal structure of anti-uPAR Fab 8B12 | | Descriptor: | anti-uPAR antibody, heavy chain, light chain | | Authors: | Zhao, B, Yuan, C, Luo, Z, Huang, M. | | Deposit date: | 2014-07-08 | | Release date: | 2015-02-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Stabilizing a flexible interdomain hinge region harboring the SMB binding site drives uPAR into its closed conformation.
J.Mol.Biol., 427, 2015
|
|
7WOK
 
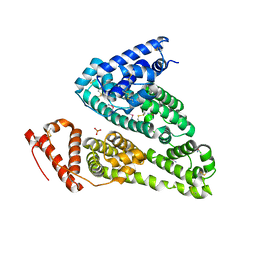 | | Crystal structure of HSA soaked with cisplatin for one week | | Descriptor: | Albumin, Cisplatin, PHOSPHATE ION | | Authors: | Chen, S.L, Yuan, C, Jiang, L.G, Luo, Z.P, Huang, M.D. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic analysis of interaction between cisplatin and human serum albumin: Effect of fatty acid.
Int.J.Biol.Macromol., 216, 2022
|
|
7WOJ
 
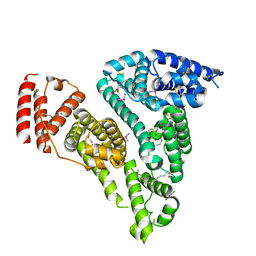 | | Crystal structure of HSA-Myr complex soaked with cisplatin for one week | | Descriptor: | Albumin, Cisplatin, MYRISTIC ACID | | Authors: | Chen, S.L, Yuan, C, Jiang, L.G, Luo, Z.P, Huang, M.D. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystallographic analysis of interaction between cisplatin and human serum albumin: Effect of fatty acid.
Int.J.Biol.Macromol., 216, 2022
|
|
4QTI
 
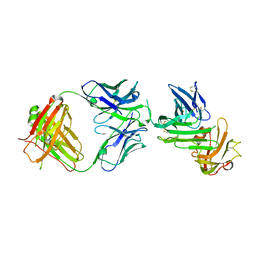 | | Crystal structure of human uPAR in complex with anti-uPAR Fab 8B12 | | Descriptor: | Urokinase plasminogen activator surface receptor, anti-uPAR antibody, heavy chain, ... | | Authors: | Zhao, B, Yuan, C, Luo, Z, Huang, M. | | Deposit date: | 2014-07-08 | | Release date: | 2015-02-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Stabilizing a flexible interdomain hinge region harboring the SMB binding site drives uPAR into its closed conformation.
J.Mol.Biol., 427, 2015
|
|
3HNY
 
 | |
1DC2
 
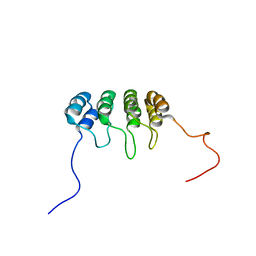 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, 20 STRUCTURES | | Descriptor: | CYCLIN-DEPENDENT KINASE 4 INHIBITOR A (P16INK4A) | | Authors: | Byeon, I.-J.L, Li, J, Yuan, C, Tsai, M.-D. | | Deposit date: | 1999-11-04 | | Release date: | 1999-12-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor INK4: refinement of p16INK4A structure and determination of p15INK4B structure by comparative modeling and NMR data.
Protein Sci., 9, 2000
|
|
1LXC
 
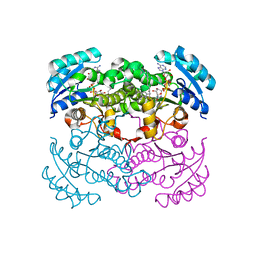 | | Crystal Structure of E. Coli Enoyl Reductase-NAD+ with a Bound Acrylamide Inhibitor | | Descriptor: | 3-(6-AMINOPYRIDIN-3-YL)-N-METHYL-N-[(1-METHYL-1H-INDOL-2-YL)METHYL]ACRYLAMIDE, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Miller, W.H, Seefeld, M.A, Newlander, K.A, Uzinskas, I.N, Burgess, W.J, Heerding, D.A, Yuan, C.C.K, Head, M.S, Payne, D.J, Rittenhouse, S.F, Moore, T.D, Pearson, S.C, Dewolf, V, Berry, W.E, Keller, P.M, Polizzi, B.J, Qiu, X, Janson, C.A, Huffman, W.F. | | Deposit date: | 2002-06-05 | | Release date: | 2002-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of aminopyridine-based inhibitors of bacterial enoyl-ACP reductase (FabI).
J.Med.Chem., 45, 2002
|
|
3HNB
 
 | |
3HOB
 
 | |
3G7N
 
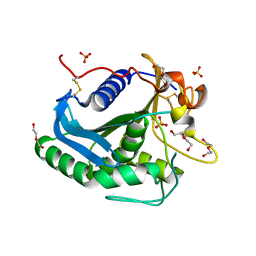 | | Crystal Structure of a Triacylglycerol Lipase from Penicillium Expansum at 1.3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lipase, PENTAETHYLENE GLYCOL, ... | | Authors: | Bian, C.B, Yuan, C, Chen, L.Q, Edward, J.M, Lin, L, Jiang, L.G, Huang, Z.X, Huang, M.D. | | Deposit date: | 2009-02-10 | | Release date: | 2010-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a triacylglycerol lipase from Penicillium expansum at 1.3 A determined by sulfur SAD
Proteins, 78, 2010
|
|
1A7A
 
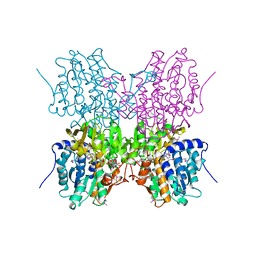 | | STRUCTURE OF HUMAN PLACENTAL S-ADENOSYLHOMOCYSTEINE HYDROLASE: DETERMINATION OF A 30 SELENIUM ATOM SUBSTRUCTURE FROM DATA AT A SINGLE WAVELENGTH | | Descriptor: | (1'R,2'S)-9-(2-HYDROXY-3'-KETO-CYCLOPENTEN-1-YL)ADENINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-ADENOSYLHOMOCYSTEINE HYDROLASE | | Authors: | Turner, M.A, Yuan, C.-S, Borchardt, R.T, Hershfield, M.S, Smith, G.D, Howell, P.L. | | Deposit date: | 1998-03-10 | | Release date: | 1999-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure determination of selenomethionyl S-adenosylhomocysteine hydrolase using data at a single wavelength.
Nat.Struct.Biol., 5, 1998
|
|
3M61
 
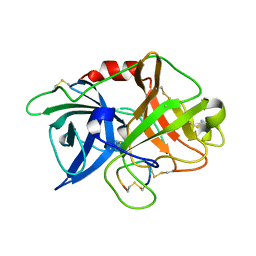 | | Crystal structure of complex of urokinase and a upain-1 variant(W3A) in pH4.6 condition | | Descriptor: | Urokinase-type plasminogen activator, upain-1 W3A | | Authors: | Jiang, L, Yuan, C, Wind, T, Andreasen, P.A, Chen, L, Meehan, E.J, Huang, M. | | Deposit date: | 2010-03-15 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of complex of urokinase and a upain-1 variant(W3A) in pH4.6 condition
TO BE PUBLISHED
|
|
7F6L
 
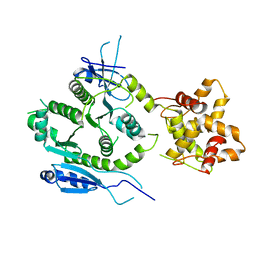 | | Crystal structure of human MUS81-EME2 complex | | Descriptor: | Crossover junction endonuclease MUS81, Probable crossover junction endonuclease EME2 | | Authors: | Hua, Z.K, Zhang, D.P, Yuan, C, Lin, Z.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the human MUS81-EME2 complex.
Structure, 30, 2022
|
|
3MWI
 
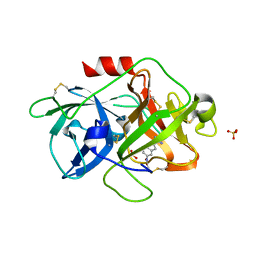 | | The complex crystal Structure of Urokianse and 5-nitro-1H-indole-2-amidine | | Descriptor: | 5-nitro-1H-indole-2-carboximidamide, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Jiang, L.G, Yu, H.Y, Yuan, C, Huang, Z.X, Huang, M.D. | | Deposit date: | 2010-05-06 | | Release date: | 2011-06-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The complex crystal Structure of Urokianse and 5-nitro-1H-indole-2-amidine
To be Published
|
|
3MHW
 
 | | The complex crystal Structure of Urokianse and 2-Aminobenzothiazole | | Descriptor: | 1,3-benzothiazol-2-amine, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Jiang, L.-G, Yuan, C, Chen, L.-Q, Huang, M.-D. | | Deposit date: | 2010-04-09 | | Release date: | 2010-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structures of 2-Aminobenzothiazole-based Inhibitors in Complexes with Urokinase-type Plasminogen Activator
CHIN.J.STRUCT.CHEM., 28, 2009
|
|
