5W0P
 
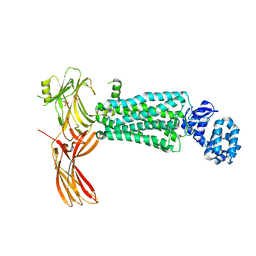 | | Crystal structure of rhodopsin bound to visual arrestin determined by X-ray free electron laser | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endolysin,Rhodopsin,S-arrestin | | Authors: | Zhou, X.E, He, Y, de Waal, P.W, Gao, X, Kang, Y, Van Eps, N, Yin, Y, Pal, K, Goswami, D, White, T.A, Barty, A, Latorraca, N.R, Chapman, H.N, Hubbell, W.L, Dror, R.O, Stevens, R.C, Cherezov, V, Gurevich, V.V, Griffin, P.R, Ernst, O.P, Melcher, K, Xu, H.E. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.013 Å) | | Cite: | Identification of Phosphorylation Codes for Arrestin Recruitment by G Protein-Coupled Receptors.
Cell, 170, 2017
|
|
4Q2E
 
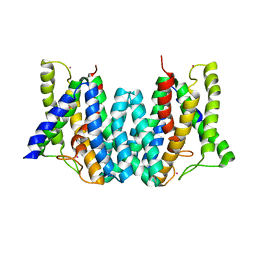 | | CRYSTAL STRUCTURE OF AN INTRAMEMBRANE CDP-DAG SYNTHETASE CENTRAL FOR PHOSPHOLIPID BIOSYNTHESIS (S200C/S258C, active mutant) | | Descriptor: | MAGNESIUM ION, MERCURY (II) ION, POTASSIUM ION, ... | | Authors: | Liu, X, Yin, Y, Wu, J, Liu, Z. | | Deposit date: | 2014-04-08 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and mechanism of an intramembrane liponucleotide synthetase central for phospholipid biosynthesis
Nat Commun, 5, 2014
|
|
4Q2G
 
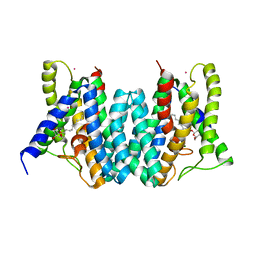 | | CRYSTAL STRUCTURE OF AN INTRAMEMBRANE CDP-DAG SYNTHETASE CENTRAL FOR PHOSPHOLIPID BIOSYNTHESIS (S200C/S223C, inactive mutant) | | Descriptor: | MAGNESIUM ION, MERCURY (II) ION, Phosphatidate cytidylyltransferase, ... | | Authors: | Liu, X, Yin, Y, Wu, J, Liu, Z. | | Deposit date: | 2014-04-08 | | Release date: | 2014-07-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and mechanism of an intramembrane liponucleotide synthetase central for phospholipid biosynthesis
Nat Commun, 5, 2014
|
|
5EDN
 
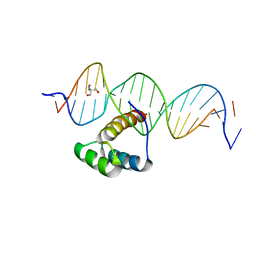 | | Structure of HOXB13-DNA(TCG) complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (5'-D(P*GP*GP*AP*CP*CP*TP*CP*GP*TP*AP*AP*AP*AP*CP*AP*CP*AP*AP*C)-3'), DNA (5'-D(P*GP*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*CP*GP*AP*GP*GP*TP*CP*C)-3'), ... | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2015-10-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Two distinct DNA sequences recognized by transcription factors represent enthalpy and entropy optima.
Elife, 7, 2018
|
|
5EEA
 
 | | Structure of HOXB13-DNA(CAA) complex | | Descriptor: | DNA (5'-D(P*GP*GP*AP*CP*CP*CP*AP*AP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*TP*TP*GP*GP*GP*TP*CP*C)-3'), Homeobox protein Hox-B13 | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2015-10-22 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Two distinct DNA sequences recognized by transcription factors represent enthalpy and entropy optima.
Elife, 7, 2018
|
|
8IJ1
 
 | | Protomer 1 and 2 of the asymmetry trimer of the Cul2-Rbx1-EloBC-FEM1B ubiquitin ligase complex | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, Elongin-B, ... | | Authors: | Dai, Z, Liang, L, Yin, Y.X. | | Deposit date: | 2023-02-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insights into the ubiquitylation strategy of the oligomeric CRL2 FEM1B E3 ubiquitin ligase.
Embo J., 43, 2024
|
|
5TDH
 
 | | The crystal structure of the dominant negative mutant G protein alpha(i)-1-beta-1-gamma-2 G203A/A326S | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, P, Jia, M.-Z, Zhou, X.E, de Waal, P.W, Dickson, B.M, Liu, B, Hou, L, Yin, Y.-T, Kang, Y.-Y, Shi, Y, Melcher, K, Xu, H.E, Jiang, Y. | | Deposit date: | 2016-09-19 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structural basis of the dominant negative phenotype of the G alpha i1 beta 1 gamma 2 G203A/A326S heterotrimer
Acta Pharmacol.Sin., 37, 2016
|
|
6IWR
 
 | | Crystal structure of GalNAc-T7 with UDP, GalNAc and Mn2+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MANGANESE (II) ION, N-acetylgalactosaminyltransferase 7, ... | | Authors: | Yu, C, Yin, Y.X. | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structural basis of carbohydrate transfer activity of UDP-GalNAc: Polypeptide N-acetylgalactosaminyltransferase 7.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
6IWQ
 
 | | Crystal structure of GalNAc-T7 with Mn2+ | | Descriptor: | MANGANESE (II) ION, N-acetylgalactosaminyltransferase 7 | | Authors: | Yu, C, Yin, Y.X. | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of carbohydrate transfer activity of UDP-GalNAc: Polypeptide N-acetylgalactosaminyltransferase 7.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
6IG4
 
 | | Structure of mitochondrial CDP-DAG synthase Tam41, delta 74 | | Descriptor: | PLATINUM (II) ION, Phosphatidate cytidylyltransferase, mitochondrial, ... | | Authors: | Jiao, H.Z, Yin, Y, Liu, Z.F. | | Deposit date: | 2018-09-23 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Structures of the Mitochondrial CDP-DAG Synthase Tam41 Suggest a Potential Lipid Substrate Pathway from Membrane to the Active Site.
Structure, 27, 2019
|
|
6IG2
 
 | | Structure of mitochondrial CDP-DAG synthase Tam41 complexed with CTP, delta 74, F240A | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, Phosphatidate cytidylyltransferase, mitochondrial | | Authors: | Jiao, H.Z, Yin, Y, Liu, Z.F. | | Deposit date: | 2018-09-23 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.882 Å) | | Cite: | Structures of the Mitochondrial CDP-DAG Synthase Tam41 Suggest a Potential Lipid Substrate Pathway from Membrane to the Active Site.
Structure, 27, 2019
|
|
6INE
 
 | | Crystal Structure of human ASH1L-MRG15 complex | | Descriptor: | GLYCEROL, Histone-lysine N-methyltransferase ASH1L, Mortality factor 4-like protein 1, ... | | Authors: | Hou, P, Huang, C, Liu, C.P, Yu, T, Yin, Y, Zhu, B, Xu, R.M. | | Deposit date: | 2018-10-25 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into Stimulation of Ash1L's H3K36 Methyltransferase Activity through Mrg15 Binding.
Structure, 27, 2019
|
|
8WPJ
 
 | |
8WPH
 
 | |
8WPI
 
 | |
8WPQ
 
 | |
8WPS
 
 | |
8WPO
 
 | | Anabaena McyI R166A with prebound NAD and citrate | | Descriptor: | CITRATE ANION, McyI, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Wang, X, Yin, Y, Duan, Y, Liu, L. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Anabaena McyI with prebound NAD and soaked NADP
To Be Published
|
|
8WPR
 
 | | Anabaena McyI R166A with prebound NAD and malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, GLYCEROL, McyI, ... | | Authors: | Wang, X, Yin, Y, Duan, Y, Liu, L. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anabaena McyI with prebound NAD and soaked NADP
To Be Published
|
|
6KTO
 
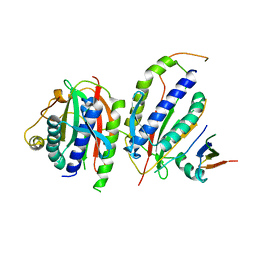 | | Crystal structure of human SHLD3-C-REV7-O-REV7-SHLD2 complex | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, Shieldin complex subunit 2, Shieldin complex subunit 3 | | Authors: | Liang, L, Yin, Y. | | Deposit date: | 2019-08-28 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4497633 Å) | | Cite: | Molecular basis for assembly of the shieldin complex and its implications for NHEJ.
Nat Commun, 11, 2020
|
|
7XK8
 
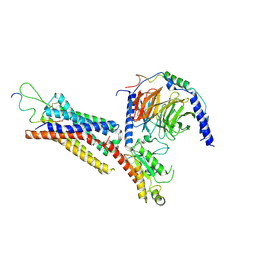 | | Cryo-EM structure of the Neuromedin U receptor 2 (NMUR2) in complex with G Protein and its endogeneous Peptide-Agonist NMU25 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, W, Wenru, Z, Mu, W, Minmin, L, Shutian, C, Tingting, T, Gisela, S, Holger, W, Albert, B, Cuiying, Y, Xiaojing, C, Han, S, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand recognition and activation of neuromedin U receptor 2.
Nat Commun, 13, 2022
|
|
6TWR
 
 | |
4UD7
 
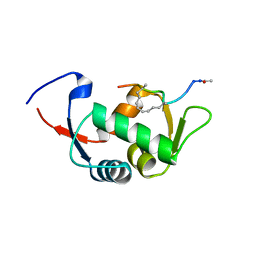 | | Structure of the stapled peptide YS-02 bound to MDM2 | | Descriptor: | MDM2, YS-02 | | Authors: | Tan, Y.S, Reeks, J, Brown, C.J, Jennings, C.E, Eapen, R.S, Tng, Q.S, Thean, D, Ying, Y.T, Gago, F.J.F, Lane, D.P, Noble, M.E.M, Verma, C. | | Deposit date: | 2014-12-08 | | Release date: | 2016-01-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Benzene Probes in Molecular Dynamics Simulations Reveal Novel Binding Sites for Ligand Design.
J Phys Chem Lett, 7, 2016
|
|
4UE1
 
 | | Structure of the stapled peptide YS-01 bound to MDM2 | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE MDM2, YS-01 | | Authors: | Tan, Y.S, Reeks, J, Brown, C.J, Jennings, C.E, Eapen, R.S, Tng, Q.S, Thean, D, Ying, Y.T, Gago, F.J.F, Lane, D.P, Noble, M.E.M, Verma, C. | | Deposit date: | 2014-12-14 | | Release date: | 2016-01-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Benzene Probes in Molecular Dynamics Simulations Reveal Novel Binding Sites for Ligand Design.
J Phys Chem Lett, 7, 2016
|
|
