2QFC
 
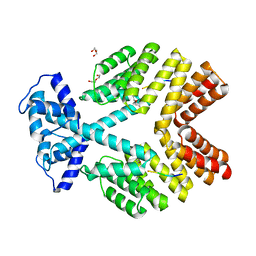 | | Crystal Structure of Bacillus thuringiensis PlcR complexed with PapR | | Descriptor: | C-terminus pentapeptide from PapR protein, DI(HYDROXYETHYL)ETHER, PlcR protein | | Authors: | Declerck, N, Chaix, D, Rugani, N, Hoh, F, Arold, S.T. | | Deposit date: | 2007-06-27 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of PlcR: Insights into virulence regulation and evolution of quorum sensing in Gram-positive bacteria
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1TLV
 
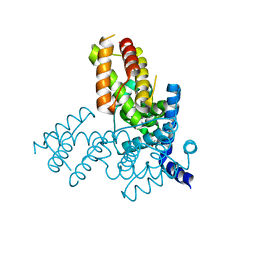 | | Structure of the native and inactive LicT PRD from B. subtilis | | Descriptor: | Transcription antiterminator licT | | Authors: | Graille, M, Zhou, C.-Z, Receveur-Brechot, V, Collinet, B, Declerck, N, van Tilbeurgh, H. | | Deposit date: | 2004-06-10 | | Release date: | 2005-02-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Activation of the LicT Transcriptional Antiterminator Involves a Domain Swing/Lock Mechanism Provoking Massive Structural Changes
J.Biol.Chem., 280, 2005
|
|
1OB0
 
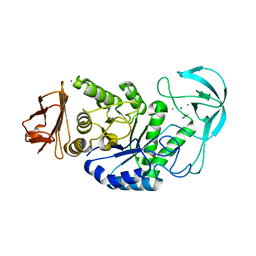 | | Kinetic stabilization of Bacillus licheniformis alpha-amylase through introduction of hydrophobic residues at the surface | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Machius, M, Declerck, N, Huber, R, Wiegand, G. | | Deposit date: | 2003-01-21 | | Release date: | 2003-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Kinetic Stabilization of Bacillus Licheniformis Alpha-Amylase Through Introduction of Hydrophobic Residues at the Surface
J.Biol.Chem., 278, 2003
|
|
6TWR
 
 | |
1L1C
 
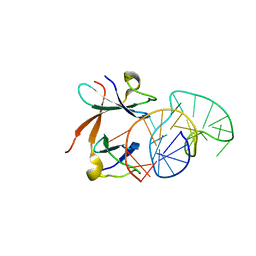 | | Structure of the LicT Bacterial Antiterminator Protein in Complex with its RNA Target | | Descriptor: | Transcription antiterminator licT, licT mRNA antiterminator hairpin | | Authors: | Yang, Y, Declerck, N, Manival, X, Aymerich, S, Kochoyan, M. | | Deposit date: | 2002-02-15 | | Release date: | 2002-03-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the LicT-RNA antitermination complex: CAT clamping RAT.
EMBO J., 21, 2002
|
|
1BLI
 
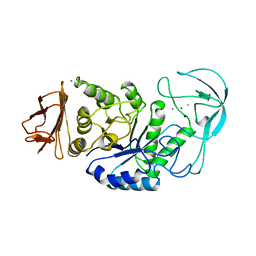 | | BACILLUS LICHENIFORMIS ALPHA-AMYLASE | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Machius, M, Declerck, N, Huber, R, Wiegand, G. | | Deposit date: | 1998-01-07 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Activation of Bacillus licheniformis alpha-amylase through a disorder-->order transition of the substrate-binding site mediated by a calcium-sodium-calcium metal triad.
Structure, 6, 1998
|
|
1H99
 
 | |
3FWR
 
 | |
3FWS
 
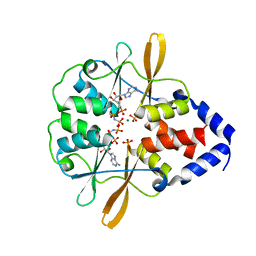 | | Crystal Structure of the CBS domains from the Bacillus subtilis CcpN repressor complexed with AppNp, phosphate and magnesium ions | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chaix, D, Arold, S, Hoh, F, Declerck, N. | | Deposit date: | 2009-01-19 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Ligand recognition by the energy sensor domain of the CcpN repressor
To be Published
|
|
3FV6
 
 | |
