8H5D
 
 | |
7Z6P
 
 | |
4WY5
 
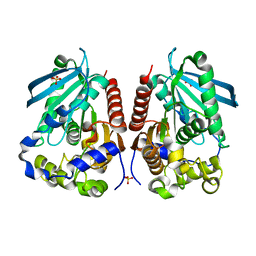 | | Structural analysis of two fungal esterases from Rhizomucor miehei explaining their substrate specificity | | 分子名称: | Esterase, SULFATE ION | | 著者 | Qin, Z, Yang, S, Duan, X, Yan, Q, Jiang, Z. | | 登録日 | 2014-11-15 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Structural insights into the substrate specificity of two esterases from the thermophilic Rhizomucor miehei
J.Lipid Res., 56, 2015
|
|
4WY8
 
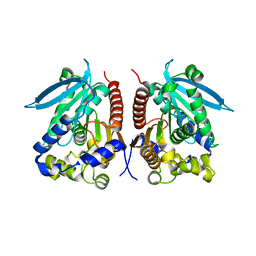 | | Structural analysis of two fungal esterases from Rhizomucor miehei explaining their substrate specificity | | 分子名称: | esterase | | 著者 | Qin, Z, Yang, S, Duan, X, Yan, Q, Jiang, Z. | | 登録日 | 2014-11-16 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Structural insights into the substrate specificity of two esterases from the thermophilic Rhizomucor miehei
J.Lipid Res., 56, 2015
|
|
8DD4
 
 | | PI 3-kinase alpha with nanobody 3-142 | | 分子名称: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | 登録日 | 2022-06-17 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD8
 
 | | PI 3-kinase alpha with nanobody 3-142, crosslinked with DSG | | 分子名称: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | 登録日 | 2022-06-17 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DCP
 
 | | PI 3-kinase alpha with nanobody 3-126 | | 分子名称: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | 登録日 | 2022-06-17 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.41 Å) | | 主引用文献 | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DCX
 
 | | PI 3-kinase alpha with nanobody 3-159 | | 分子名称: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | 登録日 | 2022-06-17 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5H9X
 
 | | Crystal structure of GH family 64 laminaripentaose-producing beta-1,3-glucanase from Paenibacillus barengoltzii | | 分子名称: | beta-1,3-glucanase | | 著者 | Zhen, Q, Yan, Q, Yang, S, Jiang, Z, You, X. | | 登録日 | 2015-12-29 | | 公開日 | 2017-02-15 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | The recognition mechanism of triple-helical beta-1,3-glucan by a beta-1,3-glucanase
Chem. Commun. (Camb.), 53, 2017
|
|
5H9Y
 
 | | Crystal structure of GH family 64 laminaripentaose-producing beta-1,3-glucanase from Paenibacillus barengoltzii complexed with laminarihexaose. | | 分子名称: | L(+)-TARTARIC ACID, beta-1,3-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | 著者 | Zhen, Q, Yan, Q, Yang, S, Jiang, Z, You, X. | | 登録日 | 2015-12-29 | | 公開日 | 2017-02-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.969 Å) | | 主引用文献 | The recognition mechanism of triple-helical beta-1,3-glucan by a beta-1,3-glucanase
Chem. Commun. (Camb.), 53, 2017
|
|
5KZF
 
 | |
7MCR
 
 | | Human Apex/Ref1 homodimer formed under oxidative condition | | 分子名称: | DNA-(apurinic or apyrimidinic site) endonuclease, mitochondrial, MAGNESIUM ION | | 著者 | Nam, Y.W, Yang, S. | | 登録日 | 2021-04-02 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The Development of Novel Apurinic/Aprymidinic Endonuclease/Redox-factor 1 Inhibitors for the Treatment of Human Melanoma
To Be Published
|
|
7MEV
 
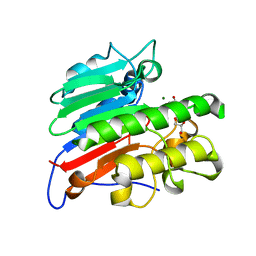 | | Human Apex/Ref1 monomer with C138A mutation | | 分子名称: | DNA-(apurinic or apyrimidinic site) endonuclease, mitochondrial, GLYCEROL, ... | | 著者 | Nam, Y.W, Yang, S. | | 登録日 | 2021-04-07 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The Development of Novel Apurinic/Aprymidinic Endonuclease/Redox-factor 1 Inhibitors for the Treatment of Human Melanoma
To Be Published
|
|
7MYO
 
 | | Cryo-EM structure of p110alpha in complex with p85alpha inhibited by BYL-719 | | 分子名称: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | 登録日 | 2021-05-21 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.92 Å) | | 主引用文献 | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MYN
 
 | | Cryo-EM Structure of p110alpha in complex with p85alpha | | 分子名称: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | 登録日 | 2021-05-21 | | 公開日 | 2021-11-10 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.79 Å) | | 主引用文献 | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7USD
 
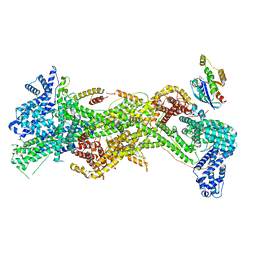 | | Cryo-EM structure of D-site Rac1-bound WAVE Regulatory Complex | | 分子名称: | Abl interactor 2, Cytoplasmic FMR1-interacting protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Ding, B, Yang, S, Chen, B, Chowdhury, S. | | 登録日 | 2022-04-25 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structures reveal a key mechanism of WAVE regulatory complex activation by Rac1 GTPase.
Nat Commun, 13, 2022
|
|
7USC
 
 | | Cryo-EM structure of WAVE Regulatory Complex | | 分子名称: | Abl interactor 2, Cytoplasmic FMR1-interacting protein 1, Nck-associated protein 1, ... | | 著者 | Ding, B, Yang, S, Chen, B, Chowdhury, S. | | 登録日 | 2022-04-25 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structures reveal a key mechanism of WAVE regulatory complex activation by Rac1 GTPase.
Nat Commun, 13, 2022
|
|
7USE
 
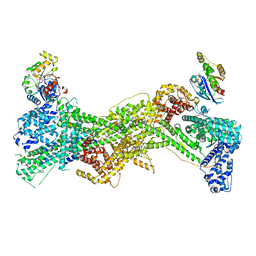 | | Cryo-EM structure of WAVE regulatory complex with Rac1 bound on both A and D site | | 分子名称: | Abl interactor 2, Cytoplasmic FMR1-interacting protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Ding, B, Yang, S, Chen, B, Chowdhury, S. | | 登録日 | 2022-04-25 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structures reveal a key mechanism of WAVE regulatory complex activation by Rac1 GTPase.
Nat Commun, 13, 2022
|
|
4ZM6
 
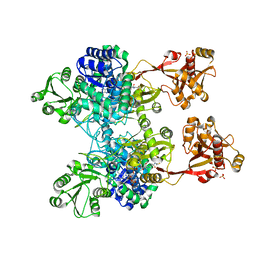 | | A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase | | 分子名称: | ACETYL COENZYME *A, N-acetyl-beta-D glucosaminidase, SULFATE ION | | 著者 | Qin, Z, Xiao, Y, Yang, X, Jiang, Z, Yang, S, Mesters, J.R. | | 登録日 | 2015-05-02 | | 公開日 | 2015-12-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain glycoside hydrolase family 3 beta-N-acetylglucosaminidase
Sci Rep, 5, 2015
|
|
7TY6
 
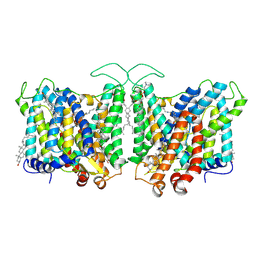 | | Cryo-EM structure of human Anion Exchanger 1 bound to 4,4'-Diisothiocyanatodihydrostilbene-2,2'-Disulfonic Acid (H2DIDS) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,4'-Diisothiocyano-2,2'-stilbenedisulfonic acid, ... | | 著者 | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | 登録日 | 2022-02-11 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TY4
 
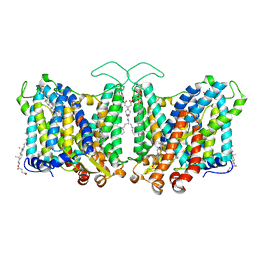 | | Cryo-EM structure of human Anion Exchanger 1 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, ... | | 著者 | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | 登録日 | 2022-02-11 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TY8
 
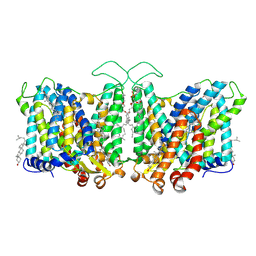 | | Cryo-EM structure of human Anion Exchanger 1 bound to Niflumic Acid | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[3-(TRIFLUOROMETHYL)PHENYL]AMINO}NICOTINIC ACID, ... | | 著者 | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | 登録日 | 2022-02-11 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TYA
 
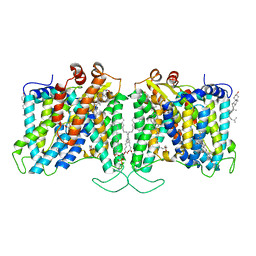 | | Cryo-EM structure of human Anion Exchanger 1 modified with Diethyl Pyrocarbonate (DEPC) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, ... | | 著者 | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | 登録日 | 2022-02-11 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TY7
 
 | | Cryo-EM structure of human Anion Exchanger 1 bound to Bicarbonate | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, ... | | 著者 | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | 登録日 | 2022-02-11 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4FBG
 
 | | Crystal structure of Treponema denticola trans-2-enoyl-CoA reductase in complex with NAD | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative reductase TDE_0597 | | 著者 | Hu, K, Zhao, M, Zhang, T, Yang, S, Ding, J. | | 登録日 | 2012-05-23 | | 公開日 | 2012-11-28 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (3.02 Å) | | 主引用文献 | Structures of trans-2-enoyl-CoA reductases from Clostridium acetobutylicum and Treponema denticola: insights into the substrate specificity and the catalytic mechanism
Biochem.J., 449, 2013
|
|
