4UX5
 
 | | Structure of DNA complex of PCG2 | | Descriptor: | 5'-D(*CP*AP*AP*TP*GP*AP*CP*GP*CP*GP*TP*AP*AP*GP)-3', 5'-D(*CP*TP*TP*AP*CP*GP*CP*GP*TP*CP*AP*TP*TP*GP)-3', TRANSCRIPTION FACTOR MBP1 | | Authors: | Liu, J, Huang, J, Zhao, Y, Liu, H, Wang, D, Yang, J, Zhao, W, Taylor, I.A, Peng, Y. | | Deposit date: | 2014-08-19 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of DNA Recognition by Pcg2 Reveals a Novel DNA Binding Mode for Winged Helix-Turn-Helix Domains.
Nucleic Acids Res., 43, 2015
|
|
8SSU
 
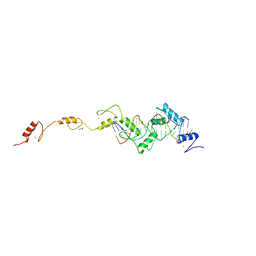 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 19mer DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (19-MER) Strand I, DNA (19-MER) Strand II, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
4OCZ
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with 1-(1-isobutyrylpiperidin-4-yl)-3-(4-(trifluoromethyl)phenyl)urea | | Descriptor: | 1-[1-(2-methylpropanoyl)piperidin-4-yl]-3-[4-(trifluoromethyl)phenyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Lee, K.S.S, Liu, J, Wagner, K.M, Pakhomova, S, Dong, H, Morriseau, C, Fu, S.H, Yang, J, Wang, P, Ulu, A, Mate, C, Nguyen, L, Wullf, H, Eldin, M.L, Mara, A.A, Newcomer, M.E, Zeldin, D.C, Hammock, B.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy.
J.Med.Chem., 57, 2014
|
|
4OD0
 
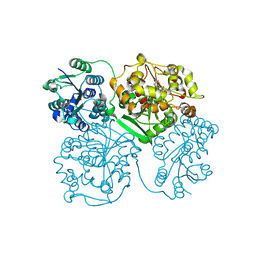 | | Crystal structure of human soluble epoxide hydrolase complexed with 1-(1-propanoylpiperidin-4-yl)-3-[4-(trifluoromethoxy)phenyl]urea | | Descriptor: | 1-(1-propanoylpiperidin-4-yl)-3-[4-(trifluoromethoxy)phenyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Lee, K.S.S, Liu, J, Wagner, K.M, Pakhomova, S, Dong, H, Morisseau, C, Fu, S.H, Yang, J, Wang, P, Ulu, A, Mate, C, Nguyen, L, Wullf, H, Eldin, M.L, Mara, A.A, Newcomer, M.E, Zeldin, D.C, Hammock, B.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy.
J.Med.Chem., 57, 2014
|
|
6LGW
 
 | | Structure of Rabies virus glycoprotein in complex with neutralizing antibody 523-11 at acidic pH | | Descriptor: | Glycoprotein, scFv 523-11 | | Authors: | Yang, F.L, Lin, S, Ye, F, Yang, J, Qi, J.X, Chen, Z.J, Lin, X, Wang, J.C, Yue, D, Cheng, Y.W, Chen, Z.M, Chen, H, You, Y, Zhang, Z.L, Yang, Y, Yang, M, Sun, H.L, Li, Y.H, Cao, Y, Yang, S.Y, Wei, Y.Q, Gao, G.F, Lu, G.W. | | Deposit date: | 2019-12-06 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9037 Å) | | Cite: | Structural Analysis of Rabies Virus Glycoprotein Reveals pH-Dependent Conformational Changes and Interactions with a Neutralizing Antibody.
Cell Host Microbe, 27, 2020
|
|
4UI9
 
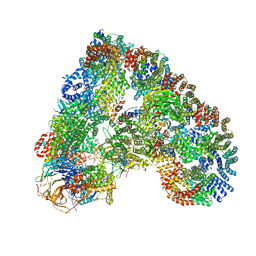 | | Atomic structure of the human Anaphase-Promoting Complex | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Chang, L, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2015-03-27 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Atomic Structure of the Apc and its Mechanism of Protein Ubiquitination
Nature, 522, 2015
|
|
2YPT
 
 | | Crystal structure of the human nuclear membrane zinc metalloprotease ZMPSTE24 mutant (E336A) in complex with a synthetic CSIM tetrapeptide from the C-terminus of prelamin A | | Descriptor: | CAAX PRENYL PROTEASE 1 HOMOLOG, PRELAMIN-A/C, ZINC ION | | Authors: | Pike, A.C.W, Dong, Y.Y, Quigley, A, Dong, L, Savitsky, P, Cooper, C.D.O, Chaikuad, A, Goubin, S, Shrestha, L, Li, Q, Mukhopadhyay, S, Yang, J, Xia, X, Shintre, C.A, Barr, A.J, Berridge, G, Chalk, R, Bray, J.E, von Delft, F, Bullock, A, Bountra, C, Arrowsmith, C.H, Edwards, A, Burgess-Brown, N, Carpenter, E.P. | | Deposit date: | 2012-11-01 | | Release date: | 2012-12-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Structural Basis of Zmpste24-Dependent Laminopathies.
Science, 339, 2013
|
|
6RPG
 
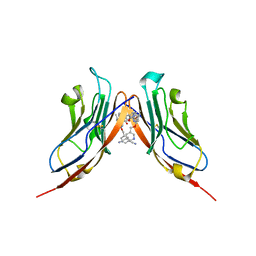 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with inhibitor | | Descriptor: | Programmed cell death 1 ligand 1, ~{N}-[2-[[4-[[3-[3-[[4-[(2-acetamidoethylamino)methyl]-5-[(5-cyanopyridin-3-yl)methoxy]-2-methyl-phenoxy]methyl]-2-methyl-phenyl]-2-methyl-phenyl]methoxy]-2-[(5-cyanopyridin-3-yl)methoxy]-5-methyl-phenyl]methylamino]ethyl]ethanamide | | Authors: | Magiera-Mularz, K, Basu, S, Yang, J, Xu, B, Skalniak, L, Musielak, B, Kholodovych, V, Holak, T.A, Hu, L. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, Synthesis, Evaluation, and Structural Studies ofC2-Symmetric Small Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
6TGB
 
 | | CryoEM structure of the binary DOCK2-ELMO1 complex | | Descriptor: | Dedicator of cytokinesis protein 2, Engulfment and cell motility protein 1 | | Authors: | Chang, L, Yang, J, Chang, J.H, Zhang, Z, Boland, A, McLaughlin, S.H, Abu-Thuraia, A, Killoran, R.C, Smith, M.J, Cote, J.F, Barford, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
7QE7
 
 | | High-resolution structure of the Anaphase-promoting complex/cyclosome (APC/C) bound to co-activator Cdh1 | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Hoefler, A, Yu, J, Chang, L, Zhang, Z, Yang, J, Boland, A, Barford, D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-01-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-resolution structure of the Anaphase-promoting complex (APC/C) bound to co-activator Cdh1
To Be Published
|
|
7N17
 
 | | Structure of TAX-4_R421W apo open state | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
7N15
 
 | | Structure of TAX-4_R421W w/cGMP open state | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CYCLIC GUANOSINE MONOPHOSPHATE, ... | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
7N16
 
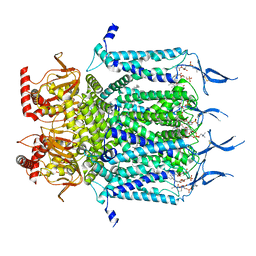 | | Structure of TAX-4_R421W apo closed state | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, SODIUM ION | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
7ON1
 
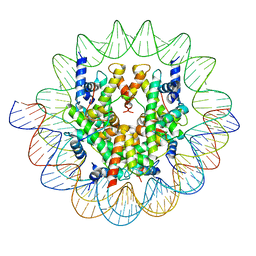 | | Cenp-A nucleosome in complex with Cenp-C | | Descriptor: | BJ4_G0006610.mRNA.1.CDS.1, BJ4_G0007000.mRNA.1.CDS.1, DNA (123-MER), ... | | Authors: | Yan, K, Yang, J, Zhang, Z, Barford, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cenp-A nucleosome in complex with Cenp-C
To Be Published
|
|
3X21
 
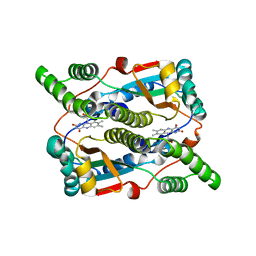 | | Crystal structure of Escherichia coli nitroreductase NfsB mutant T41L/N71S/F124W | | Descriptor: | FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Bai, J, Yang, J, Zhou, Y, Yang, Q. | | Deposit date: | 2014-12-06 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Altering the regioselectivity of a nitroreductase in the synthesis of arylhydroxylamines by structure-based engineering.
Chembiochem, 16, 2015
|
|
3X22
 
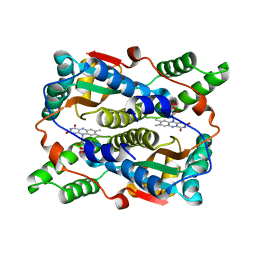 | | Crystal structure of Escherichia coli nitroreductase NfsB mutant N71S/F123A/F124W | | Descriptor: | FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Bai, J, Yang, J, Zhou, Y, Yang, Q. | | Deposit date: | 2014-12-06 | | Release date: | 2015-11-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis of Escherichia coli nitroreductase NfsB triple mutants engineered for improved activity and regioselectivity toward the prodrug CB1954
PROCESS BIOCHEM, 50, 2015
|
|
6JUI
 
 | | The atypical Myb-like protein Cdc5 contains two distinct nucleic acid-binding surfaces | | Descriptor: | Pre-mRNA-splicing factor CEF1 | | Authors: | Wang, C, Li, G, Li, M, Yang, J, Liu, J. | | Deposit date: | 2019-04-14 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Two distinct nucleic acid binding surfaces of Cdc5 regulate development.
Biochem.J., 476, 2019
|
|
7YGW
 
 | | Crystal structure of the Zn2+-bound EFhd1/Swiprosin-2 | | Descriptor: | EF-hand domain-containing protein D1, GLYCEROL, ZINC ION | | Authors: | Mun, S.A, Park, J, Kang, J.Y, Park, T, Jin, M, Yang, J, Eom, S.H. | | Deposit date: | 2022-07-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural and biochemical insights into Zn 2+ -bound EF-hand proteins, EFhd1 and EFhd2.
Iucrj, 10, 2023
|
|
7YGY
 
 | | Crystal structure of the Zn2+-bound EFhd2/Swiprosin-1 | | Descriptor: | EF-hand domain-containing protein D2, ZINC ION | | Authors: | Mun, S.A, Park, J, Kang, J.Y, Park, T, Jin, M, Yang, J, Eom, S.H. | | Deposit date: | 2022-07-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical insights into Zn 2+ -bound EF-hand proteins, EFhd1 and EFhd2.
Iucrj, 10, 2023
|
|
8EU3
 
 | | Cryo-EM structure of cGMP bound human CNGA3/CNGB3 channel in GDN, transition state 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel alpha-3, ... | | Authors: | Hu, Z, Zheng, X, Yang, J. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Conformational trajectory of allosteric gating of the human cone photoreceptor cyclic nucleotide-gated channel.
Nat Commun, 14, 2023
|
|
8ETP
 
 | | Cryo-EM structure of cGMP bound closed state of human CNGA3/CNGB3 channel in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel alpha-3, ... | | Authors: | Hu, Z, Zheng, X, Yang, J. | | Deposit date: | 2022-10-17 | | Release date: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Conformational trajectory of allosteric gating of the human cone photoreceptor cyclic nucleotide-gated channel.
Nat Commun, 14, 2023
|
|
8EUC
 
 | | Cryo-EM structure of cGMP bound human CNGA3/CNGB3 channel in GDN, transition state 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel alpha-3, ... | | Authors: | Hu, Z, Zheng, X, Yang, J. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Conformational trajectory of allosteric gating of the human cone photoreceptor cyclic nucleotide-gated channel.
Nat Commun, 14, 2023
|
|
8EVA
 
 | |
8EVB
 
 | |
8EV9
 
 | |
