1F9P
 
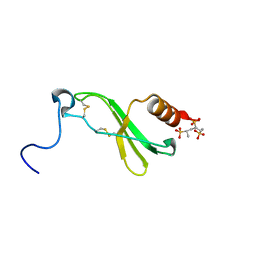 | | CRYSTAL STRUCTURE OF CONNECTIVE TISSUE ACTIVATING PEPTIDE-III(CTAP-III) COMPLEXED WITH POLYVINYLSULFONIC ACID | | Descriptor: | CONNECTIVE TISSUE ACTIVATING PEPTIDE-III, ETHANESULFONIC ACID | | Authors: | Yang, J, Faulk, T, Aster, R, Visentin, G, Edwards, B, Castor, C. | | Deposit date: | 2000-07-11 | | Release date: | 2003-08-26 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of the CXC Chemokine, Connective Tissue Activating Peptide-III, Complexed with the Heparin Analogue, Polyvinylsulfonic Acid
To be Published
|
|
1F9S
 
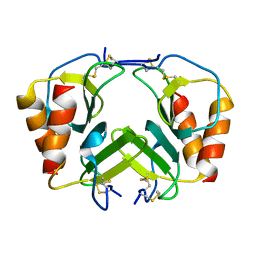 | | CRYSTAL STRUCTURE OF PLATELET FACTOR 4 MUTANT 2 | | Descriptor: | PLATELET FACTOR 4 | | Authors: | Yang, J, Doyle, M, Faulk, T, Visentin, G, Aster, R, Edwards, B. | | Deposit date: | 2000-07-11 | | Release date: | 2003-08-26 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure Comparison of Two Platelet Factor 4 Mutants with the Wild-type Reveals the Epitopes for the Heparin-induced Thrombocytopenia Antibodies
To be Published
|
|
1GWZ
 
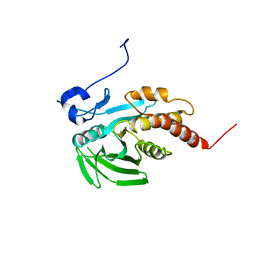 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF THE PROTEIN TYROSINE PHOSPHATASE SHP-1 | | Descriptor: | SHP-1 | | Authors: | Yang, J, Liang, X, Niu, T, Meng, W, Zhao, Z, Zhou, G.W. | | Deposit date: | 1998-08-22 | | Release date: | 1999-08-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the catalytic domain of protein-tyrosine phosphatase SHP-1.
J.Biol.Chem., 273, 1998
|
|
6JHJ
 
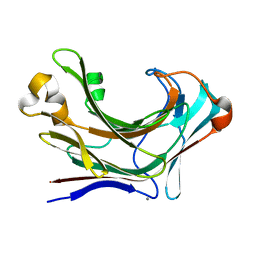 | | Structure of Marine bacterial laminarinase mutant-E135A | | Descriptor: | CALCIUM ION, LamCAT | | Authors: | Yang, J, Xu, Y, Miyakawa, T, Tanokura, M, Long, L. | | Deposit date: | 2019-02-18 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
6JH5
 
 | | Structure of Marine bacterial laminarinase | | Descriptor: | CALCIUM ION, LamCAT | | Authors: | Yang, J, Xu, Y, Miyakawa, T, Ru, L, Tanokura, M, Long, L. | | Deposit date: | 2019-02-17 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
6JIA
 
 | | Marine bacterial laminarinase mutant E135A complex with laminaritetraose | | Descriptor: | CALCIUM ION, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose, laminarinase | | Authors: | Yang, J, Xu, Y, Miyakawa, T, Tanokura, M, Long, L. | | Deposit date: | 2019-02-20 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
1RDQ
 
 | | Hydrolysis of ATP in the crystal of Y204A mutant of cAMP-dependent protein kinase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yang, J, Ten Eyck, L.F, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2003-11-05 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal Structure of a cAMP-dependent Protein Kinase Mutant at 1.26A: New Insights into the Catalytic Mechanism.
J.Mol.Biol., 336, 2004
|
|
8D9X
 
 | | Cryo-EM structure of human DELE1 in oligomeric form | | Descriptor: | Maltodextrin-binding protein,DAP3-binding cell death enhancer 1 short form | | Authors: | Yang, J, Lander, G.C. | | Deposit date: | 2022-06-11 | | Release date: | 2023-06-14 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | DELE1 oligomerization promotes integrated stress response activation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8J0P
 
 | | Chitin binding SusD-like protein AqSusD from a marine Bacteroidetes | | Descriptor: | Chitin binding SusD-like protein | | Authors: | Yang, J. | | Deposit date: | 2023-04-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights of a SusD-like protein in marine Bacteroidetes bacteria reveal the molecular basis for chitin recognition and acquisition.
Febs J., 291, 2024
|
|
1RE5
 
 | | Crystal structure of 3-carboxy-cis,cis-muconate lactonizing enzyme from Pseudomonas putida | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-carboxy-cis,cis-muconate cycloisomerase, CITRIC ACID | | Authors: | Yang, J, Wang, Y, Woolridge, E.M, Petsko, G.A, Kozarich, J.W, Ringe, D. | | Deposit date: | 2003-11-06 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of 3-Carboxy-cis,cis-muconate Lactonizing Enzyme from Pseudomonas putida, a Fumarase Class II Type Cycloisomerase: Enzyme Evolution in Parallel Pathways.
Biochemistry, 43, 2004
|
|
1RXT
 
 | | Crystal structure of human myristoyl-CoA:protein N-myristoyltransferase. | | Descriptor: | COBALT (II) ION, Glycylpeptide N-tetradecanoyltransferase 1, SULFATE ION | | Authors: | Yang, J, Wang, Y, Frey, G, Abeles, R.H, Petsko, G.A, Ringe, D. | | Deposit date: | 2003-12-18 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human myristoyl-CoA:protein N-myristoyltransferase
To be Published
|
|
1O6K
 
 | | Structure of activated form of PKB kinase domain S474D with GSK3 peptide and AMP-PNP | | Descriptor: | GLYCOGEN SYNTHASE KINASE-3 BETA, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Yang, J, Cron, P, Good, V.M, Thompson, V, Hemmings, B.A, Barford, D. | | Deposit date: | 2002-10-08 | | Release date: | 2002-11-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of an Activated Akt/Protein Kinase B Ternary Complex with Gsk-3 Peptide and AMP-Pnp
Nat.Struct.Biol., 9, 2002
|
|
1O6L
 
 | | Crystal structure of an activated Akt/protein kinase B (PKB-PIF chimera) ternary complex with AMP-PNP and GSK3 peptide | | Descriptor: | GLYCOGEN SYNTHASE KINASE-3 BETA, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Yang, J, Cron, P, Good, V.M, Thompson, V, Hemmings, B.A, Barford, D. | | Deposit date: | 2002-10-08 | | Release date: | 2002-11-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of an Activated Akt/Protein Kinase B Ternary Complex with Gsk-3 Peptide and AMP-Pnp
Nat.Struct.Biol., 9, 2002
|
|
6PZP
 
 | | Crystal structure of caspase-1 in complex with VX-765 | | Descriptor: | Caspase-1, N-(4-amino-3-chlorobenzene-1-carbonyl)-3-methyl-L-valyl-N-[(2S)-1-carboxy-3-oxopropan-2-yl]-L-prolinamide | | Authors: | Yang, J, Liu, Z, Xiao, T.S. | | Deposit date: | 2019-08-01 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of caspase-1 in complex with VX-765
To Be Published
|
|
2KNC
 
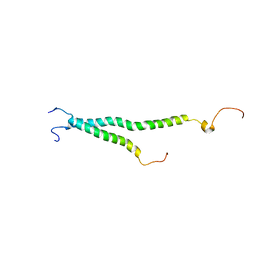 | | Platelet integrin ALFAIIB-BETA3 transmembrane-cytoplasmic heterocomplex | | Descriptor: | Integrin alpha-IIb, Integrin beta-3 | | Authors: | Yang, J, Ma, Y, Page, R.C, Misra, S, Plow, E.F, Qin, J. | | Deposit date: | 2009-08-20 | | Release date: | 2009-09-29 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of an integrin alphaIIb beta3 transmembrane-cytoplasmic heterocomplex provides insight into integrin activation.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3QAM
 
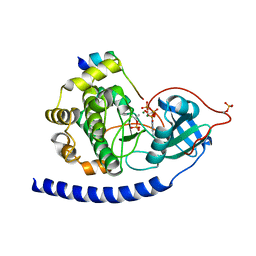 | | Crystal Structure of Glu208Ala mutant of catalytic subunit of cAMP-dependent protein kinase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein kinase inhibitor, ... | | Authors: | Yang, J, Wu, J, Steichen, J, Taylor, S.S. | | Deposit date: | 2011-01-11 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A conserved Glu-Arg salt bridge connects coevolved motifs that define the eukaryotic protein kinase fold.
J.Mol.Biol., 415, 2012
|
|
3QAL
 
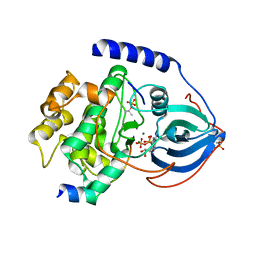 | | Crystal Structure of Arg280Ala mutant of Catalytic subunit of cAMP-dependent Protein Kinase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein kinase inhibitor, ... | | Authors: | Yang, J, Wu, J, Steichen, J, Taylor, S.S. | | Deposit date: | 2011-01-11 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A conserved Glu-Arg salt bridge connects coevolved motifs that define the eukaryotic protein kinase fold.
J.Mol.Biol., 415, 2012
|
|
3R4L
 
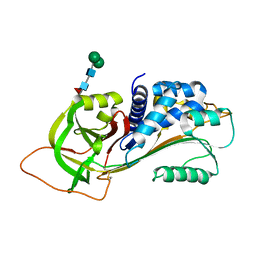 | | Human very long half life Plasminogen Activator Inhibitor type-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plasminogen activator inhibitor 1, beta-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yang, J, Zheng, H, Han, Q, Skrzypczak-Jankun, E, Jankun, J. | | Deposit date: | 2011-03-17 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Remarkable extension of PAI-1 half-life surprisingly brings no changes to its structure.
Int.J.Mol.Med., 29, 2012
|
|
3PYY
 
 | | Discovery and Characterization of a Cell-Permeable, Small-molecule c-Abl Kinase Activator that Binds to the Myristoyl Binding Site | | Descriptor: | (5R)-5-[3-(4-fluorophenyl)-1-phenyl-1H-pyrazol-4-yl]imidazolidine-2,4-dione, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, GLYCEROL, ... | | Authors: | Yang, J, Campobasso, N, Biju, M.P, Fisher, K, Pan, X.Q, Cottom, J, Galbraith, S, Ho, T, Zhang, H, Hong, X, Ward, P, Hofmann, G, Siegfried, B. | | Deposit date: | 2010-12-13 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and Characterization of a Cell-Permeable, Small-Molecule c-Abl Kinase Activator that Binds to the Myristoyl Binding Site.
Chem.Biol., 18, 2011
|
|
2MWN
 
 | | Talin-F3 / RIAM N-terminal Peptide complex | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1-interacting protein, Talin-1 | | Authors: | Yang, J, Zhu, L, Zhang, H, Hirbawi, J, Fukuda, K, Dwivedi, P, Liu, J, Byzova, T, Plow, E.F, Wu, J, Qin, J. | | Deposit date: | 2014-11-13 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational activation of talin by RIAM triggers integrin-mediated cell adhesion.
Nat Commun, 5, 2014
|
|
8JJB
 
 | | Crystal structure of T2R-TTL-Y61 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yang, J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure-based design and synthesis of BML284 derivatives: A novel class of colchicine-site noncovalent tubulin degradation agents.
Eur.J.Med.Chem., 268, 2024
|
|
8JJC
 
 | | Tubulin-Y62 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(6,7-dimethoxy-3,4-dihydro-1~{H}-isoquinolin-2-yl)-6-(3-methoxyphenyl)pyrimidin-2-amine, CALCIUM ION, ... | | Authors: | Yang, J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure-based design and synthesis of BML284 derivatives: A novel class of colchicine-site noncovalent tubulin degradation agents.
Eur.J.Med.Chem., 268, 2024
|
|
6LPX
 
 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with 2-oxoglutarate (2-OG) | | Descriptor: | 2-OXOGLUTARIC ACID, D-2-hydroxyglutarate dehydrogenase, mitochondrial, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6LPP
 
 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with D-2-hydroxyglutarate (D-2-HG) | | Descriptor: | (2R)-2-hydroxypentanedioic acid, D-2-hydroxyglutarate dehydrogenase, mitochondrial, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
6LPU
 
 | | Crystal structure of human D-2-hydroxyglutarate dehydrogenase in complex with L-2-hydroxyglutarate (L-2-HG) | | Descriptor: | (2S)-2-HYDROXYPENTANEDIOIC ACID, D-2-hydroxyglutarate dehydrogenase, mitochondrial, ... | | Authors: | Yang, J, Zhu, H, Ding, J. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.923 Å) | | Cite: | Structure, substrate specificity, and catalytic mechanism of human D-2-HGDH and insights into pathogenicity of disease-associated mutations.
Cell Discov, 7, 2021
|
|
