2KNC
 
 | | Platelet integrin ALFAIIB-BETA3 transmembrane-cytoplasmic heterocomplex | | Descriptor: | Integrin alpha-IIb, Integrin beta-3 | | Authors: | Yang, J, Ma, Y, Page, R.C, Misra, S, Plow, E.F, Qin, J. | | Deposit date: | 2009-08-20 | | Release date: | 2009-09-29 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of an integrin alphaIIb beta3 transmembrane-cytoplasmic heterocomplex provides insight into integrin activation.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8KFA
 
 | | Cryo-EM structure of HSV-1 gB with D48 Fab complex | | Descriptor: | D48 heavy chain, D48 light chain, Envelope glycoprotein B | | Authors: | Yang, J, Sun, C, Fang, X, Zeng, M, Liu, Z. | | Deposit date: | 2023-08-15 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | The structure of HSV-1 gB bound to a potent neutralizing antibody reveals a conservative antigenic domain across herpesviruses
hlife, 2023
|
|
8H5U
 
 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain in complex with neutralizing nanobody Nb-021 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody Nb-021, ... | | Authors: | Yang, J, Lin, S, Lu, G.W. | | Deposit date: | 2022-10-13 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Development of a bispecific nanobody conjugate broadly neutralizes diverse SARS-CoV-2 variants and structural basis for its broad neutralization.
Plos Pathog., 19, 2023
|
|
8H5T
 
 | |
5HM2
 
 | |
2MWN
 
 | | Talin-F3 / RIAM N-terminal Peptide complex | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1-interacting protein, Talin-1 | | Authors: | Yang, J, Zhu, L, Zhang, H, Hirbawi, J, Fukuda, K, Dwivedi, P, Liu, J, Byzova, T, Plow, E.F, Wu, J, Qin, J. | | Deposit date: | 2014-11-13 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational activation of talin by RIAM triggers integrin-mediated cell adhesion.
Nat Commun, 5, 2014
|
|
8IL3
 
 | | Cryo-EM structure of CD38 in complex with FTL004 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, Heavy chain, Light chain | | Authors: | Yang, J, Wang, Y, Zhang, G. | | Deposit date: | 2023-03-01 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | FTL004, an anti-CD38 mAb with negligible RBC binding and enhanced pro-apoptotic activity, is a novel candidate for treatments of multiple myeloma and non-Hodgkin lymphoma.
J Hematol Oncol, 15, 2022
|
|
8W75
 
 | | Structure of Drosophila melanogaster L-2-hydroxyglutarate dehydrogenase | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FI05204p, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Yang, J, Chen, X, Jin, S, Ding, J. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and biochemical characterization of l-2-hydroxyglutarate dehydrogenase and its role in the pathogenesis of l-2-hydroxyglutaric aciduria.
J.Biol.Chem., 300, 2023
|
|
8W7F
 
 | | Structure of Drosophila melanogaster L-2-hydroxyglutarate dehydrogenase bound with FAD and a sulfate ion | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FI05204p, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yang, J, Chen, X, Jin, S, Ding, J. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structure and biochemical characterization of l-2-hydroxyglutarate dehydrogenase and its role in the pathogenesis of l-2-hydroxyglutaric aciduria.
J.Biol.Chem., 300, 2023
|
|
8W78
 
 | | Structure of Drosophila melanogaster L-2-hydroxyglutarate dehydrogenase in complex with FAD and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, DODECYL-BETA-D-MALTOSIDE, FI05204p, ... | | Authors: | Yang, J, Chen, X, Jin, S, Ding, J. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure and biochemical characterization of l-2-hydroxyglutarate dehydrogenase and its role in the pathogenesis of l-2-hydroxyglutaric aciduria.
J.Biol.Chem., 300, 2023
|
|
8WD0
 
 | | Crystal structure of T2R-TTL-Erianin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-methoxy-5-[2-(3,4,5-trimethoxyphenyl)ethyl]phenol, CALCIUM ION, ... | | Authors: | Yang, J. | | Deposit date: | 2023-09-14 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The cytotoxic natural compound erianin binds to colchicine site of beta-tubulin and overcomes taxane resistance
Bioorg.Chem., 150, 2024
|
|
7SXO
 
 | | Yeast Lon (PIM1) with endogenous substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Lon protease homolog, ... | | Authors: | Yang, J, Song, A.S, Wiseman, R.L, Lander, G.C. | | Deposit date: | 2021-11-24 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of hexameric yeast Lon protease (PIM1) highlights the importance of conserved structural elements.
J.Biol.Chem., 298, 2022
|
|
7W1S
 
 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain in complex with neutralizing nanobody Nb-007 | | Descriptor: | Nanobody Nb-007, Spike protein S1 | | Authors: | Yang, J, Lin, S, Sun, H.L, Lu, G.W. | | Deposit date: | 2021-11-20 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | A Potent Neutralizing Nanobody Targeting the Spike Receptor-Binding Domain of SARS-CoV-2 and the Structural Basis of Its Intimate Binding.
Front Immunol, 13, 2022
|
|
8XBD
 
 | |
8XHO
 
 | |
8XPH
 
 | | Marine Planctomycetes laminarinase PtLam | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Laminarinase PtLam | | Authors: | Yang, J. | | Deposit date: | 2024-01-03 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Marine Planctomycetes laminarinase PtLam
To Be Published
|
|
8XPK
 
 | |
8J0P
 
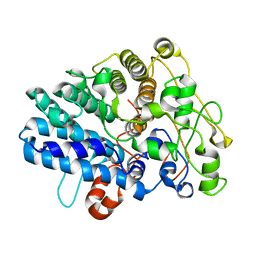 | | Chitin binding SusD-like protein AqSusD from a marine Bacteroidetes | | Descriptor: | Chitin binding SusD-like protein | | Authors: | Yang, J. | | Deposit date: | 2023-04-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights of a SusD-like protein in marine Bacteroidetes bacteria reveal the molecular basis for chitin recognition and acquisition.
Febs J., 291, 2024
|
|
2LR7
 
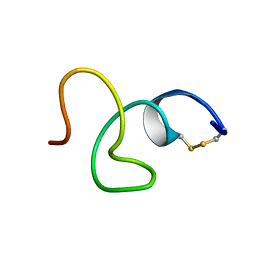 | | Cathelicidin-PY | | Descriptor: | Cathelicidin-PY | | Authors: | Yang, J. | | Deposit date: | 2012-03-27 | | Release date: | 2013-03-27 | | Method: | SOLUTION NMR | | Cite: | Structure of Cathelicidin-PY
To be Published
|
|
8XRH
 
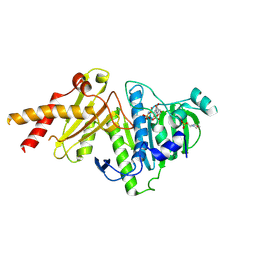 | |
8XRG
 
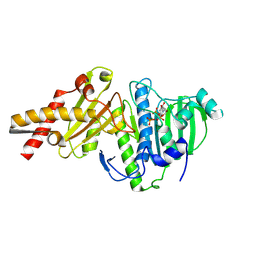 | |
8XRI
 
 | |
8XRF
 
 | |
6JHJ
 
 | | Structure of Marine bacterial laminarinase mutant-E135A | | Descriptor: | CALCIUM ION, LamCAT | | Authors: | Yang, J, Xu, Y, Miyakawa, T, Tanokura, M, Long, L. | | Deposit date: | 2019-02-18 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
6JH5
 
 | | Structure of Marine bacterial laminarinase | | Descriptor: | CALCIUM ION, LamCAT | | Authors: | Yang, J, Xu, Y, Miyakawa, T, Ru, L, Tanokura, M, Long, L. | | Deposit date: | 2019-02-17 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
