5GJJ
 
 | |
7DP8
 
 | | Crystal structure of T2R-TTL-Cevipabulin-eribulin complex | | Descriptor: | (1S,3S,6S,9S,12S,14R,16R,18S,20R,21R,22S,26R,29S,31R,32S,33R,35R,36S)-20-[(2S)-3-amino-2-hydroxypropyl]-21-methoxy-14-methyl-8,15-dimethylidene-2,19,30,34,37,39,40,41-octaoxanonacyclo[24.9.2.1~3,32~.1~3,33~.1~6,9~.1~12,16~.0~18,22~.0~29,36~.0~31,35~]hentetracontan-24-one (non-preferred name), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[2,6-bis(fluoranyl)-4-[3-(methylamino)propoxy]phenyl]-5-chloranyl-N-[(2S)-1,1,1-tris(fluoranyl)propan-2-yl]-[1,2,4]triazolo[1,5-a]pyrimidin-7-amine, ... | | Authors: | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | Deposit date: | 2020-12-18 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.446 Å) | | Cite: | Cevipabulin-tubulin complex reveals a novel agent binding site on alpha-tubulin with tubulin degradation effect.
Sci Adv, 7, 2021
|
|
3NX1
 
 | | Crystal structure of Enterobacter sp. Px6-4 Ferulic Acid Decarboxylase | | Descriptor: | Ferulic acid decarboxylase | | Authors: | Gu, W, Yang, J.K, Lou, Z.Y, Meng, Z.H, Zhang, K.-Q. | | Deposit date: | 2010-07-12 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Enzymatic Activity for the Ferulic Acid Decarboxylase (FADase) from Enterobacter sp. Px6-4
Plos One, 6, 2011
|
|
4JNE
 
 | | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Hsp70 CHAPERONE DnaK, ... | | Authors: | Qi, R, Sarbeng, E.B, Liu, Q, Le, K.Q, Xu, X, Xu, H, Yang, J, Wong, J.L, Vorvis, C, Hendrickson, W.A, Zhou, L, Liu, Q. | | Deposit date: | 2013-03-15 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3NX2
 
 | | Enterobacter sp. Px6-4 Ferulic Acid Decarboxylase in complex with substrate analogues | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, Ferulic acid decarboxylase | | Authors: | Gu, W, Yang, J.K, Lou, Z.Y, Meng, Z.H, Zhang, K.-Q. | | Deposit date: | 2010-07-12 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis of Enzymatic Activity for the Ferulic Acid Decarboxylase (FADase) from Enterobacter sp. Px6-4
Plos One, 6, 2011
|
|
4MRN
 
 | | Structure of a bacterial Atm1-family ABC transporter | | Descriptor: | ABC transporter related protein, LAURYL DIMETHYLAMINE-N-OXIDE, PHOSPHATE ION | | Authors: | Lee, J.Y, Yang, J.G, Zhitnitsky, D, Lewinson, O, Rees, D.C. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for heavy metal detoxification by an Atm1-type ABC exporter.
Science, 343, 2014
|
|
4MRS
 
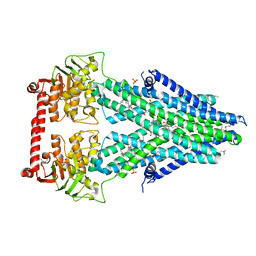 | | Structure of a bacterial Atm1-family ABC transporter | | Descriptor: | ABC transporter related protein, LAURYL DIMETHYLAMINE-N-OXIDE, OXIDIZED GLUTATHIONE DISULFIDE, ... | | Authors: | Lee, J.Y, Yang, J.G, Zhitnitsky, D, Lewinson, O, Rees, D.C. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-19 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for heavy metal detoxification by an Atm1-type ABC exporter.
Science, 343, 2014
|
|
4MRR
 
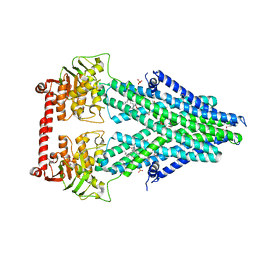 | | Structure of a bacterial Atm1-family ABC transporter | | Descriptor: | ABC transporter related protein, LAURYL DIMETHYLAMINE-N-OXIDE, PHOSPHATE ION, ... | | Authors: | Lee, J.Y, Yang, J.G, Zhitnitsky, D, Lewinson, O, Rees, D.C. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-19 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural basis for heavy metal detoxification by an Atm1-type ABC exporter.
Science, 343, 2014
|
|
2QUR
 
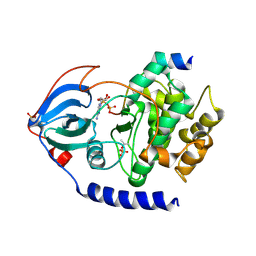 | | Crystal Structure of F327A/K285P Mutant of cAMP-dependent Protein Kinase | | Descriptor: | 20-mer fragment from cAMP-dependent protein kinase inhibitor alpha, ADENOSINE-5'-DIPHOSPHATE, cAMP-dependent protein kinase, ... | | Authors: | Taylor, S.S, Yang, J, Wu, J. | | Deposit date: | 2007-08-06 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Contribution of non-catalytic core residues to activity and regulation in protein kinase A.
J.Biol.Chem., 284, 2009
|
|
5YDG
 
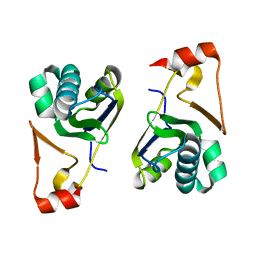 | | Crystal structure of the Arabidopsis thaliana chloroplast RNA editing factors 2(MORF2) | | Descriptor: | Multiple organellar RNA editing factor 2, chloroplastic | | Authors: | Wang, X, Yang, J.Y, Wang, Y.L, Gao, Y.S. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Crystal structure of the chloroplast RNA editing factor MORF2
Biochem. Biophys. Res. Commun., 495, 2018
|
|
6JFK
 
 | | GDP bound Mitofusin2 (MFN2) | | Descriptor: | CITRIC ACID, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Li, Y.J, Cao, Y.L, Feng, J.X, Qi, Y.B, Meng, S.X, Yang, J.F, Zhong, Y.T, Kang, S.S, Chen, X.X, Lan, L, Luo, L, Yu, B, Chen, S.D, Chan, D.C, Hu, J.J, Gao, S. | | Deposit date: | 2019-02-10 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural insights of human mitofusin-2 into mitochondrial fusion and CMT2A onset.
Nat Commun, 10, 2019
|
|
6JFL
 
 | | Nucleotide-free Mitofusin2 (MFN2) | | Descriptor: | CALCIUM ION, GLYCEROL, Mitofusin-2,cDNA FLJ57997, ... | | Authors: | Li, Y.J, Cao, Y.L, Feng, J.X, Qi, Y.B, Meng, S.X, Yang, J.F, Zhong, Y.T, Kang, S.S, Chen, X.X, Lan, L, Luo, L, Yu, B, Chen, S.D, Chan, D.C, Hu, J.J, Gao, S. | | Deposit date: | 2019-02-10 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structural insights of human mitofusin-2 into mitochondrial fusion and CMT2A onset.
Nat Commun, 10, 2019
|
|
6JFM
 
 | | Mitofusin2 (MFN2)_T111D | | Descriptor: | ACETATE ION, CALCIUM ION, Mitofusin-2,Mitofusin-2 | | Authors: | Li, Y.J, Cao, Y.L, Feng, J.X, Qi, Y.B, Meng, S.X, Yang, J.F, Zhong, Y.T, Kang, S.S, Chen, X.X, Lan, L, Luo, L, Yu, B, Chen, S.D, Chan, D.C, Hu, J.J, Gao, S. | | Deposit date: | 2019-02-10 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural insights of human mitofusin-2 into mitochondrial fusion and CMT2A onset.
Nat Commun, 10, 2019
|
|
5ZXH
 
 | | The structure of MT189-tubulin complex | | Descriptor: | 2-(6-fluoro-3-{[(4-methoxyphenyl)methyl]amino}imidazo[1,2-a]pyridin-2-yl)phenol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Li, Z.P, Wang, Y.X, Meng, T, Yang, J.L, Chen, Q. | | Deposit date: | 2018-05-21 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of MT189-Tubulin Complex Provides Insights into Drug Design
Lett.Drug Des.Discovery, 2019
|
|
2LBM
 
 | |
3QCA
 
 | |
8PKP
 
 | | Cryo-EM structure of the apo Anaphase-promoting complex/cyclosome (APC/C) at 3.2 Angstrom resolution | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Hoefler, A, Yu, J, Chang, L, Zhang, Z, Yang, J, Boland, A, Barford, D. | | Deposit date: | 2023-06-27 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution structure of the Anaphase-promoting complex (APC/C) bound to co-activator Cdh1
To Be Published
|
|
7BV3
 
 | | Crystal structure of a ugt transferase from Siraitia grosvenorii in complex with UDP | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2020-04-09 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Near-perfect control of the regioselective glucosylation enabled by rational design of glycosyltransferases
Green Synth Catal, 2021
|
|
4JNF
 
 | | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP | | Descriptor: | Hsp70 CHAPERONE DnaK | | Authors: | Qi, R, Sarbeng, E.B, Liu, Q, Le, K.Q, Xu, X, Xu, H, Yang, J, Wong, J.L, Vorvis, C, Hendrickson, W.A, Zhou, L, Liu, Q. | | Deposit date: | 2013-03-15 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.621 Å) | | Cite: | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP.
Nat.Struct.Mol.Biol., 20, 2013
|
|
8OVW
 
 | | Cryo-EM structure of CBF1-CCAN bound topologically to centromeric DNA | | Descriptor: | C0N3 DNA, Centromere-binding protein 1, Inner kinetochore subunit AME1, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
8OW0
 
 | | Cryo-EM structure of CBF1-CCAN bound topologically to a centromeric CENP-A nucleosome | | Descriptor: | C0N3 DNA, Centromere-binding protein 1, Histone H2A.1, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
8OW1
 
 | | Cryo-EM structure of the yeast Inner kinetochore bound to a CENP-A nucleosome. | | Descriptor: | C0N3, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
2KQC
 
 | | Second PBZ domain of human APLF protein | | Descriptor: | Aprataxin and PNK-like factor, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Brockmann, C, Yang, J. | | Deposit date: | 2009-11-04 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose).
Nat.Struct.Mol.Biol., 17, 2010
|
|
2KQB
 
 | | First PBZ domain of human APLF protein | | Descriptor: | Aprataxin and PNK-like factor, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Brockmann, C, Yang, J. | | Deposit date: | 2009-11-04 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose).
Nat.Struct.Mol.Biol., 17, 2010
|
|
2HQE
 
 | | Crystal structure of human P100 Tudor domain: Large fragment | | Descriptor: | P100 Co-activator tudor domain | | Authors: | Shah, N, Zhao, M, Cheng, C, Xu, H, Yang, J, Silvennoinen, O, Liu, Z.J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-18 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a large fragment of the Human P100 Tudor Domain
To be Published
|
|
