8W8S
 
 | | Cryo-EM structure of the AA14-bound GPR101 complex | | Descriptor: | 1-(4-methylpyridin-2-yl)-3-[3-(trifluoromethyl)phenyl]thiourea, Probable G-protein coupled receptor 101 | | Authors: | Sun, J.P, Yu, X, Gao, N, Yang, F, Wang, J.Y, Yang, Z, Guan, Y, Wang, G.P. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
8W8Q
 
 | | Cryo-EM structure of the GPR101-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Sun, J.P, Gao, N, Yu, X, Wang, G.P, Yang, F, Wang, J.Y, Yang, Z, Guan, Y. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
6N3G
 
 | | Crystal structure of histone lysine methyltransferase SmyD2 in complex with polyethylene glycol | | Descriptor: | DODECAETHYLENE GLYCOL, ETHANOL, N-lysine methyltransferase SMYD2, ... | | Authors: | Perry, E, Spellmon, N, Brunzelle, J, Yang, Z. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structure of histone lysine methyltransferase SmyD2 in complex with polyethylene glycol
To be Published
|
|
3JZI
 
 | | Crystal structure of biotin carboxylase from E. Coli in complex with benzimidazole series | | Descriptor: | 7-amino-2-[(2-chlorobenzyl)amino]-1-{[(1S,2S)-2-hydroxycycloheptyl]methyl}-1H-benzimidazole-5-carboxamide, Biotin carboxylase | | Authors: | Cheng, C, Shipps, G.W, Yang, Z, Sun, B, Kawahata, N, Soucy, K, Soriano, A, Orth, P, Xiao, L, Mann, P, Black, T. | | Deposit date: | 2009-09-23 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery and optimization of antibacterial AccC inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
7YKS
 
 | |
7YKR
 
 | |
5AF0
 
 | | MAEL domain from Bombyx mori Maelstrom | | Descriptor: | MAELSTROM, ZINC ION | | Authors: | Chen, K, Campbell, E, Pandey, R.R, Yang, Z, McCarthy, A.A, Pillai, R.S. | | Deposit date: | 2015-01-13 | | Release date: | 2015-04-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Metazoan Maelstrom is an RNA-Binding Protein that Has Evolved from an Ancient Nuclease Active in Protists.
RNA, 21, 2015
|
|
1U2Z
 
 | | Crystal structure of histone K79 methyltransferase Dot1p from yeast | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Sawada, K, Yang, Z, Horton, J.R, Collins, R.E, Zhang, X, Cheng, X. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase
J.Biol.Chem., 279, 2004
|
|
3DV2
 
 | | Crystal Structure of nicotinic acid mononucleotide adenylyltransferase from Bacillus anthracis | | Descriptor: | Nicotinate (Nicotinamide) nucleotide adenylyltransferase, SULFATE ION | | Authors: | Lu, S, Smith, C.D, Yang, Z, Pruett, P.S, Nagy, L, McCombs, D.P, DeLucas, L.J, Brouillette, W.J, Brouillette, C.G. | | Deposit date: | 2008-07-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of nicotinic acid mononucleotide adenylyltransferase from Bacillus anthracis.
ACTA CRYSTALLOGR.,SECT.F, 64, 2008
|
|
4Z0W
 
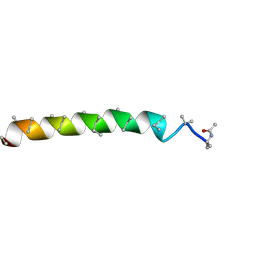 | | Peptaibol gichigamin isolated from Tolypocladium sup_5 | | Descriptor: | PEPTAIBOL GICHIGAMIN | | Authors: | Du, L, Risinger, A.L, Mitchell, C.A, Stamps, B.W, Pan, N, King, J.B, Motley, J.L, Thomas, L.M, Yang, Z, Stevenson, B.S, Mooberry, S.L, Cichewicz, R.H. | | Deposit date: | 2015-03-26 | | Release date: | 2016-03-30 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Peptaibol gichigamin isolated from Tolypocladium sup_5
TO BE PUBLISHED
|
|
4P0C
 
 | | Crystal Structure of NHERF2 PDZ1 Domain in Complex with LPA2 | | Descriptor: | CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF2/Lysophosphatidic acid receptor 2 chimeric protein, THIOCYANATE ION | | Authors: | Holcomb, J, Jiang, Y, Lu, G, Trescott, L, Brunzelle, J, Sirinupong, N, Li, C, Naren, A, Yang, Z. | | Deposit date: | 2014-02-20 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.339 Å) | | Cite: | Structural insights into PDZ-mediated interaction of NHERF2 and LPA2, a cellular event implicated in CFTR channel regulation.
Biochem.Biophys.Res.Commun., 446, 2014
|
|
3PDN
 
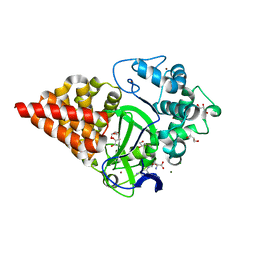 | | Crystal structure of SmyD3 in complex with methyltransferase inhibitor sinefungin | | Descriptor: | GLYCEROL, MAGNESIUM ION, SET and MYND domain-containing protein 3, ... | | Authors: | Sirinupong, N, Brunzelle, J, Yang, Z. | | Deposit date: | 2010-10-23 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights into the Autoinhibition and Posttranslational Activation of Histone Methyltransferase SmyD3.
J.Mol.Biol., 406, 2011
|
|
3HI1
 
 | | Structure of HIV-1 gp120 (core with V3) in Complex with CD4-Binding-Site Antibody F105 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F105 Heavy Chain, F105 Light Chain, ... | | Authors: | Kwon, Y.D, Chen, L, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z, Zhang, M.-Y, Arthos, J, Burton, D.R, Dimitrov, D, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-05-18 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
6MON
 
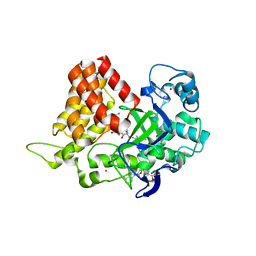 | | Crystal structure of human SMYD2 in complex with Nle-peptide inhibitor | | Descriptor: | GLYCEROL, LYS-LEU-NLE-SER-LYS-ARG-GLY, N-lysine methyltransferase SMYD2, ... | | Authors: | Spellmon, N, Cornett, E, Brunzelle, J, Rothbart, S, Yang, Z. | | Deposit date: | 2018-10-04 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | A functional proteomics platform to reveal the sequence determinants of lysine methyltransferase substrate selectivity.
Sci Adv, 4, 2018
|
|
4PQW
 
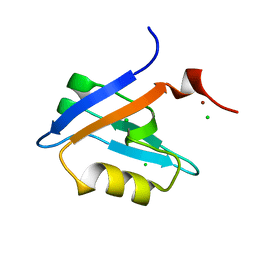 | | Crystal Structure of Phospholipase C beta 3 in Complex with PDZ1 of NHERF1 | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Jiang, Y, Wang, S, Holcomb, J, Trescott, L, Guan, X, Hou, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallographic analysis of NHERF1-PLC beta 3 interaction provides structural basis for CXCR2 signaling in pancreatic cancer.
Biochem.Biophys.Res.Commun., 446, 2014
|
|
3QWW
 
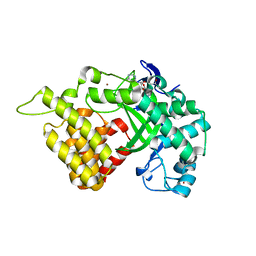 | | Crystal structure of histone lysine methyltransferase SmyD2 in complex with the methyltransferase inhibitor sinefungin | | Descriptor: | SET and MYND domain-containing protein 2, SINEFUNGIN, ZINC ION | | Authors: | Jiang, Y, Sirinupong, N, Brunzelle, J, Yang, Z. | | Deposit date: | 2011-02-28 | | Release date: | 2011-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of histone and p53 methyltransferase SmyD2 reveal a conformational flexibility of the autoinhibitory C-terminal domain.
Plos One, 6, 2011
|
|
3QWV
 
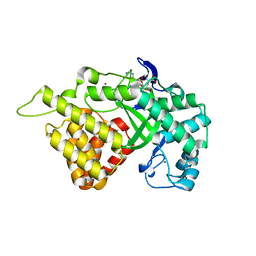 | | Crystal structure of histone lysine methyltransferase SmyD2 in complex with the cofactor product AdoHcy | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SET and MYND domain-containing protein 2, ZINC ION | | Authors: | Jiang, Y, Sirinupong, N, Brunzelle, J, Yang, Z. | | Deposit date: | 2011-02-28 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structures of histone and p53 methyltransferase SmyD2 reveal a conformational flexibility of the autoinhibitory C-terminal domain.
Plos One, 6, 2011
|
|
3N71
 
 | |
4MPA
 
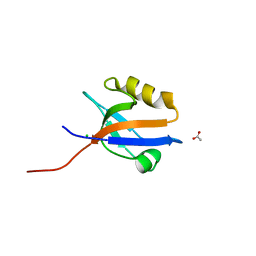 | | Crystal structure of NHERF1-CXCR2 signaling complex in P21 space group | | Descriptor: | ACETIC ACID, CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1, ... | | Authors: | Jiang, Y, Lu, G, Wu, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2013-09-12 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.097 Å) | | Cite: | New Conformational State of NHERF1-CXCR2 Signaling Complex Captured by Crystal Lattice Trapping.
Plos One, 8, 2013
|
|
1OD2
 
 | | Acetyl-CoA Carboxylase Carboxyltransferase Domain | | Descriptor: | ACETYL COENZYME *A, ACETYL-COENZYME A CARBOXYLASE, ADENINE | | Authors: | Zhang, H, Yang, Z, Shen, Y, Tong, L. | | Deposit date: | 2003-02-12 | | Release date: | 2003-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase.
Science, 299, 2003
|
|
4O6F
 
 | | Structural Basis of Estrogen Receptor Alpha Methylation Mediated by Histone Methyltransferase SmyD2 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Estrogen receptor, N-lysine methyltransferase SMYD2, ... | | Authors: | Jiang, Y, Trescott, L, Holcomb, J, Brunzelle, J, Sirinupong, N, Shi, X, Yang, Z. | | Deposit date: | 2013-12-20 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.822 Å) | | Cite: | Structural Insights into Estrogen Receptor alpha Methylation by Histone Methyltransferase SMYD2, a Cellular Event Implicated in Estrogen Signaling Regulation.
J.Mol.Biol., 426, 2014
|
|
4Q3H
 
 | | The crystal structure of NHERF1 PDZ2 CXCR2 complex revealed by the NHERF1 CXCR2 chimeric protein | | Descriptor: | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Holcomb, J, Jiang, Y, Trescott, L, Lu, G, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2014-04-11 | | Release date: | 2014-05-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.443 Å) | | Cite: | Crystal structure of the NHERF1 PDZ2 domain in complex with the chemokine receptor CXCR2 reveals probable modes of PDZ2 dimerization.
Biochem.Biophys.Res.Commun., 448, 2014
|
|
1PEG
 
 | | Structural basis for the product specificity of histone lysine methyltransferases | | Descriptor: | Histone H3, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Zhang, X, Yang, Z, Khan, S.I, Horton, J.R, Tamaru, H, Selker, E.U, Cheng, X. | | Deposit date: | 2003-05-21 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis for the product specificity of histone lysine methyltransferases
Mol.Cell, 12, 2003
|
|
8TH7
 
 | |
5TYT
 
 | | Crystal Structure of the PDZ domain of RhoGEF bound to CXCR2 C-terminal peptide | | Descriptor: | Rho guanine nucleotide exchange factor 11, C-X-C chemokine receptor type 2 chimera | | Authors: | Spellmon, N, Holcomb, J, Niu, A, Choudhary, V, Sun, X, Brunzelle, J, Li, C, Yang, Z. | | Deposit date: | 2016-11-21 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structural basis of PDZ-mediated chemokine receptor CXCR2 scaffolding by guanine nucleotide exchange factor PDZ-RhoGEF.
Biochem. Biophys. Res. Commun., 485, 2017
|
|
