1KKX
 
 | | Solution structure of the DNA-binding domain of ADR6 | | Descriptor: | Transcription regulatory protein ADR6 | | Authors: | Tu, X, Wu, J, Xu, Y, Shi, Y. | | Deposit date: | 2001-12-10 | | Release date: | 2002-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments and secondary structure of ADR6 DNA-binding domain.
J.Biomol.Nmr, 21, 2001
|
|
1SJ6
 
 | | NMR Structure and Regulated Expression in APL Cell of Human SH3BGRL3 | | Descriptor: | SH3 domain-binding glutamic acid-rich-like protein 3 | | Authors: | Xu, C, Tang, Y, Xu, Y, Wu, J, Shi, Y, Zhang, Q, Zheng, P, Du, Y. | | Deposit date: | 2004-03-03 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure and regulated expression in APL cell of human SH3BGRL3.
Febs Lett., 579, 2005
|
|
5SXU
 
 | | X-ray structure of 2-bromoethanol bound to a pentameric ligand gated ion channel (ELIC) in a desensitized state | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-BROMOETHANOL, 3-AMINOPROPANE, ... | | Authors: | Chen, Q, Kinde, M, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2016-08-10 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Alcohol Inhibition of the Pentameric Ligand-Gated Ion Channel ELIC.
Structure, 25, 2017
|
|
5SXV
 
 | | X-ray structure of 2-bromoethanol bound to a pentameric ligand gated ion channel (ELIC) in a resting state | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-BROMOETHANOL, Cys-loop ligand-gated ion channel | | Authors: | Chen, Q, Kinde, M, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2016-08-10 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Alcohol Inhibition of the Pentameric Ligand-Gated Ion Channel ELIC.
Structure, 25, 2017
|
|
8XVV
 
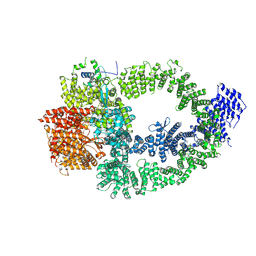 | | The TRRAP module of human NuA4/TIP60 complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Isoform 2 of E1A-binding protein p400, Isoform 2 of Transformation/transcription domain-associated protein | | Authors: | Chen, K, Wang, L, Yu, Z, Yu, J, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human TIP60 complex.
Nat Commun, 15, 2024
|
|
8XVG
 
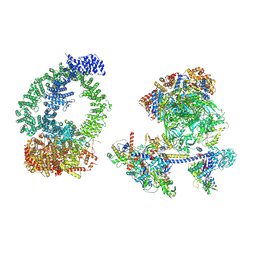 | | Structure of human NuA4/TIP60 complex | | Descriptor: | ACTB protein (Fragment), ADENOSINE-5'-DIPHOSPHATE, Actin-like protein 6A, ... | | Authors: | Chen, K, Wang, L, Yu, Z, Yu, J, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structure of the human TIP60 complex.
Nat Commun, 15, 2024
|
|
8XVT
 
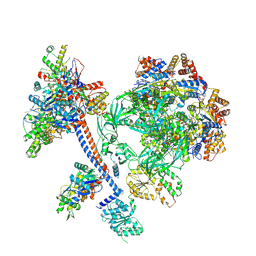 | | The core subcomplex of human NuA4/TIP60 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Chen, K, Wang, L, Yu, Z, Yu, J, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human TIP60 complex.
Nat Commun, 15, 2024
|
|
7MYO
 
 | | Cryo-EM structure of p110alpha in complex with p85alpha inhibited by BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | Deposit date: | 2021-05-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MYN
 
 | | Cryo-EM Structure of p110alpha in complex with p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | Deposit date: | 2021-05-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7YL2
 
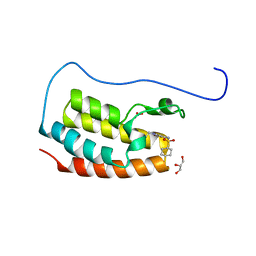 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y07004 | | Descriptor: | Bromodomain-containing protein 4, GLYCEROL, N-(1-ethyl-2-oxidanylidene-3H-indol-5-yl)cyclohexanesulfonamide, ... | | Authors: | Huang, Y, Wei, A, Dong, R, Xu, H, Zhang, C, Chen, Z, Li, J, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-07-25 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y07004
To Be Published
|
|
5V54
 
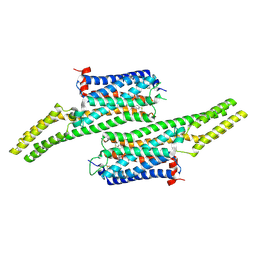 | | Crystal structure of 5-HT1B receptor in complex with methiothepin | | Descriptor: | 1-methyl-4-[(5~{S})-3-methylsulfanyl-5,6-dihydrobenzo[b][1]benzothiepin-5-yl]piperazine, 5-hydroxytryptamine receptor 1B,OB-1 fused 5-HT1b receptor,5-hydroxytryptamine receptor 1B | | Authors: | Yin, W.C, Zhou, X.E, Yang, D, de Waal, P, Wang, M.T, Dai, A, Cai, X, Huang, C.Y, Liu, P, Yin, Y, Liu, B, Caffrey, M, Melcher, K, Xu, Y, Wang, M.W, Xu, H.E, Jiang, Y. | | Deposit date: | 2017-03-13 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | A common antagonistic mechanism for class A GPCRs revealed by the structure of the human 5-HT1B serotonin receptor bound to an antagonist
Cell Discov, 2018
|
|
4LGB
 
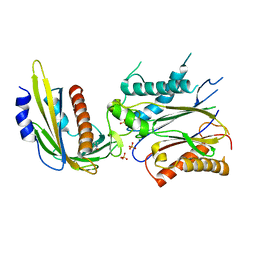 | | ABA-mimicking ligand N-(1-METHYL-2-OXO-1,2,3,4-TETRAHYDROQUINOLIN-6-YL)-1-(4-METHYLPHENYL)METHANESULFONAMIDE in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | Abscisic acid receptor PYL2, MAGNESIUM ION, N-(1-methyl-2-oxo-1,2,3,4-tetrahydroquinolin-6-yl)-1-(4-methylphenyl)methanesulfonamide, ... | | Authors: | Zhou, X.E, Gao, M, Liu, X, Zhang, Y, Xue, X, Melcher, K, Gao, P, Wang, F, Zeng, L, Zhao, Y, Zhao, Y, Deng, P, Zhong, D, Zhu, J.-K, Xu, Y, Xu, H.E. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants.
Cell Res., 23, 2013
|
|
4LG5
 
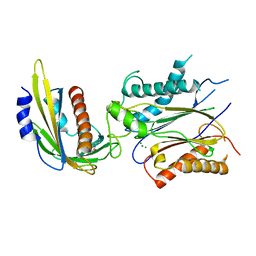 | | ABA-mimicking ligand QUINABACTIN in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | Abscisic acid receptor PYL2, MAGNESIUM ION, Protein phosphatase 2C 16, ... | | Authors: | Zhou, X.E, Gao, M, Liu, X, Zhang, Y, Xue, X, Melcher, K, Gao, P, Wang, F, Zeng, L, Zhao, Y, Zhao, Y, Deng, P, Zhong, D, Zhu, J.-K, Xu, Y, Xu, H.E. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants.
Cell Res., 23, 2013
|
|
4LGA
 
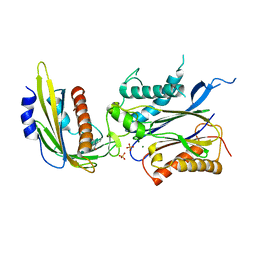 | | ABA-mimicking ligand N-(2-OXO-1-PROPYL-1,2,3,4-TETRAHYDROQUINOLIN-6-YL)-1-PHENYLMETHANESULFONAMIDE in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | Abscisic acid receptor PYL2, MAGNESIUM ION, N-(2-oxo-1-propyl-1,2,3,4-tetrahydroquinolin-6-yl)-1-phenylmethanesulfonamide, ... | | Authors: | Zhou, X.E, Gao, M, Liu, X, Zhang, Y, Xue, X, Melcher, K, Gao, P, Wang, F, Zeng, L, Zhao, Y, Zhao, Y, Deng, P, Zhong, D, Zhu, J.-K, Xu, Y, Xu, H.E. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants.
Cell Res., 23, 2013
|
|
2A7P
 
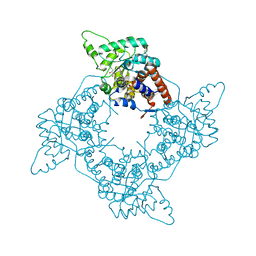 | | Crystal Structure of the G81A mutant of the Active Chimera of (S)-Mandelate Dehydrogenase in complex with its substrate 3-indolelactate | | Descriptor: | (S)-Mandelate Dehydrogenase, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-(INDOL-3-YL) LACTATE, ... | | Authors: | Sukumar, N, Xu, Y, Mitra, B, Mathews, F.S. | | Deposit date: | 2005-07-05 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the G81A mutant form of the active chimera of (S)-mandelate dehydrogenase and its complex with two of its substrates.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3PUQ
 
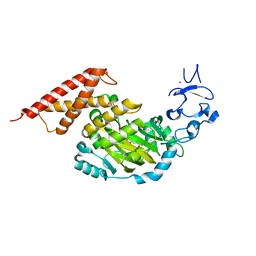 | | CEKDM7A from C.Elegans, complex with alpha-KG | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, GLYCEROL, ... | | Authors: | Yang, Y, Wang, P, Xu, W, Xu, Y. | | Deposit date: | 2010-12-06 | | Release date: | 2011-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases
Cancer Cell, 19, 2011
|
|
6HN5
 
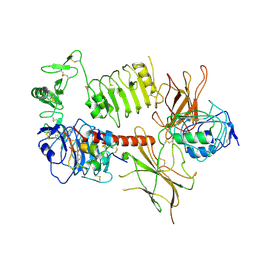 | | Leucine-zippered human insulin receptor ectodomain with single bound insulin - "upper" membrane-distal part | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin, ... | | Authors: | Weis, F, Menting, J.G, Margetts, M.B, Chan, S.J, Xu, Y, Tennagels, N, Wohlfart, P, Langer, T, Mueller, C.W, Dreyer, M.K, Lawrence, M.C. | | Deposit date: | 2018-09-14 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The signalling conformation of the insulin receptor ectodomain.
Nat Commun, 9, 2018
|
|
3PUR
 
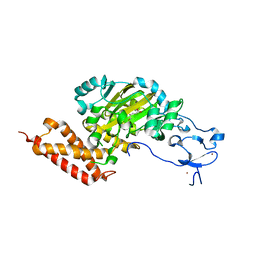 | | CEKDM7A from C.Elegans, complex with D-2-HG | | Descriptor: | (2R)-2-hydroxypentanedioic acid, FE (II) ION, Lysine-specific demethylase 7 homolog, ... | | Authors: | Yang, Y, Wang, P, Xu, W, Xu, Y. | | Deposit date: | 2010-12-06 | | Release date: | 2011-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases
Cancer Cell, 19, 2011
|
|
8J0I
 
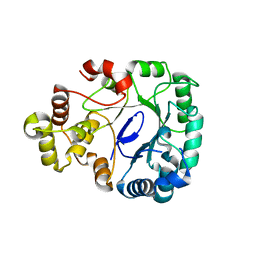 | | Aldo-keto reductase KmAKR | | Descriptor: | NADPH-dependent alpha-keto amide reductase, SODIUM ION | | Authors: | Xu, S.Y, Zhou, L, Xu, Y, Wang, Y.J, Zheng, Y.G. | | Deposit date: | 2023-04-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Aldo-keto reductase KmAKR from Kluyveromyces marxianus
To Be Published
|
|
8JWY
 
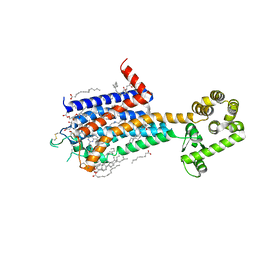 | | Crystal structure of A2AR-T4L in complex with 2-118 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-azanyl-6-[2-oxidanylidene-1-[[6-(2-oxidanylpropan-2-yl)pyridin-2-yl]methyl]pyridin-4-yl]pyrimidin-4-yl]-2-methyl-benzenecarbonitrile, Adenosine receptor A2a,Endolysin, ... | | Authors: | Weng, Y, Chen, Y, Xu, Y, Song, G. | | Deposit date: | 2023-06-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insight into the dual-antagonistic mechanism of AB928 on adenosine A 2 receptors.
Sci China Life Sci, 67, 2024
|
|
8JWZ
 
 | | Crystal structure of A2AR-T4L in complex with AB928 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-azanyl-6-[1-[[6-(2-oxidanylpropan-2-yl)pyridin-2-yl]methyl]-1,2,3-triazol-4-yl]pyrimidin-4-yl]-2-methyl-benzenecarbonitrile, Adenosine receptor A2a,Endolysin, ... | | Authors: | Weng, Y, Chen, Y, Xu, Y, Song, G. | | Deposit date: | 2023-06-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural insight into the dual-antagonistic mechanism of AB928 on adenosine A 2 receptors.
Sci China Life Sci, 67, 2024
|
|
4WQN
 
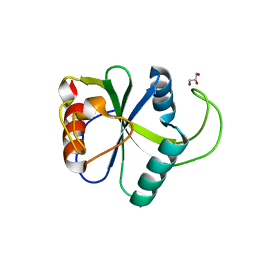 | | Crystal structure of N6-methyladenosine RNA reader YTHDF2 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, YTH domain-containing family protein 2 | | Authors: | Zhu, T, Roundtree, I.A, Wang, P, Wang, X, Wang, L, Sun, C, Tian, Y, Li, J, He, C, Xu, Y. | | Deposit date: | 2014-10-22 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine.
Cell Res., 24, 2014
|
|
6P72
 
 | | Crystal Structure of the Cedar henipavirus Attachment G Glycoprotein global domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein, ... | | Authors: | Xu, K, Nikolov, D.B, Xu, Y. | | Deposit date: | 2019-06-04 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.283 Å) | | Cite: | Structural and functional analyses reveal promiscuous and species specific use of ephrin receptors by Cedar virus.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6P7S
 
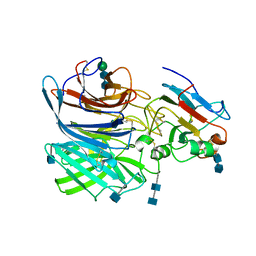 | | Crystal Structure of the Cedar henipavirus Attachment G Glycoprotein globular domain in complex with the receptor ephrin-B1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein, ... | | Authors: | Xu, K, Nikolov, D.B, Xu, Y. | | Deposit date: | 2019-06-06 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structural and functional analyses reveal promiscuous and species specific use of ephrin receptors by Cedar virus.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6P7Y
 
 | | Crystal Structure of the Cedar henipavirus Attachment G Glycoprotein globular domain in complex with the receptor ephrin-B2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein, ... | | Authors: | Xu, K, Nikolov, D.B, Xu, Y. | | Deposit date: | 2019-06-06 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.844 Å) | | Cite: | Structural and functional analyses reveal promiscuous and species specific use of ephrin receptors by Cedar virus.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
