2PV3
 
 | |
5WA5
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor XMD11-50 | | 分子名称: | 1,2-ETHANEDIOL, 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl}phenyl)amino]-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4 | | 著者 | Xu, X, Blacklow, S.C. | | 登録日 | 2017-06-24 | | 公開日 | 2018-07-04 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.172 Å) | | 主引用文献 | Structural and Atropisomeric Factors Governing the Selectivity of Pyrimido-benzodiazipinones as Inhibitors of Kinases and Bromodomains.
ACS Chem. Biol., 13, 2018
|
|
4Y2W
 
 | |
7XKA
 
 | | Structure of human beta2 adrenergic receptor bound to constrained epinephrine | | 分子名称: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Camelid Antibody Fragment, Endolysin,Beta-2 adrenergic receptor, ... | | 著者 | Xu, X, Shonberg, J, Kaindl, J, Clark, M, Stobel, A, Maul, L, Mayer, D, Hubner, H, Venkatakrishnan, A, Dror, R, Kobilka, B.K, Sunahara, R, Liu, X, Gmeiner, P. | | 登録日 | 2022-04-19 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Constrained catecholamines gain beta 2 AR selectivity through allosteric effects on pocket dynamics.
Nat Commun, 14, 2023
|
|
7XK9
 
 | | Structure of human beta2 adrenergic receptor bound to constrained isoproterenol | | 分子名称: | (5R,6R)-6-(propan-2-ylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Camelid Antibody Fragment, Endolysin,Beta-2 adrenergic receptor, ... | | 著者 | Xu, X, Shonberg, J, Kaindl, J, Clark, M, Stobel, A, Maul, L, Mayer, D, Hubner, H, Venkatakrishnan, A, Dror, R, Kobilka, B.K, Sunahara, R, Liu, X, Gmeiner, P. | | 登録日 | 2022-04-19 | | 公開日 | 2023-04-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Constrained catecholamines gain beta 2 AR selectivity through allosteric effects on pocket dynamics.
Nat Commun, 14, 2023
|
|
5M82
 
 | | Three-dimensional structure of the photoproduct state of GAF3 from Slr1393 of Synechocystis sp. PCC6803 | | 分子名称: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, PHYCOCYANOBILIN, SODIUM ION, ... | | 著者 | Xu, X.-L, Zhao, K.-H, Gaertner, W, Hoeppner, A. | | 登録日 | 2016-10-28 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Structural elements regulating the photochromicity in a cyanobacteriochrome
Proc.Natl.Acad.Sci.USA, 2020
|
|
5M85
 
 | | Three-dimensional structure of the intermediate state of GAF3 from Slr1393 of Synechocystis sp. PCC6803 | | 分子名称: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHYCOCYANOBILIN, ... | | 著者 | Xu, X.-L, Zhao, K.-H, Gaertner, W, Hoeppner, A. | | 登録日 | 2016-10-28 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural elements regulating the photochromicity in a cyanobacteriochrome
Proc.Natl.Acad.Sci.USA, 2020
|
|
4WVL
 
 | | Structure-Guided DOT1L Probe Optimization by Label-Free Ligand Displacement | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Histone-lysine N-methyltransferase, ... | | 著者 | Xu, X, Dhe-Paganon, S, Blacklow, S. | | 登録日 | 2014-11-06 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structure-Guided DOT1L Probe Optimization by Label-Free Ligand Displacement.
Acs Chem.Biol., 10, 2015
|
|
1T09
 
 | | Crystal structure of human cytosolic NADP(+)-dependent isocitrate dehydrogenase in complex NADP | | 分子名称: | Isocitrate dehydrogenase [NADP] cytoplasmic, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Xu, X, Zhao, J, Peng, B, Huang, Q, Arnold, E, Ding, J. | | 登録日 | 2004-04-08 | | 公開日 | 2004-06-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity
J.Biol.Chem., 279, 2004
|
|
1T0L
 
 | | Crystal structure of human cytosolic NADP(+)-dependent isocitrate dehydrogenase in complex with NADP, isocitrate, and calcium(2+) | | 分子名称: | CALCIUM ION, ISOCITRIC ACID, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | 著者 | Xu, X, Zhao, J, Peng, B, Huang, Q, Arnold, E, Ding, J. | | 登録日 | 2004-04-10 | | 公開日 | 2004-06-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity
J.Biol.Chem., 279, 2004
|
|
8AN5
 
 | | MenAT1 toxin-antitoxin complex (rv0078a-rv0078b) from Mycobacterium tuberculosis H37Rv | | 分子名称: | Bacterial toxin, Conserved protein | | 著者 | Xu, X, Usher, B, Gutierrez, C, Barriot, R, Arrowsmith, T.J, Han, X, Redder, P, Neyrolles, O, Blower, T.R, Genevaux, P. | | 登録日 | 2022-08-04 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | MenT nucleotidyltransferase toxins extend tRNA acceptor stems and can be inhibited by asymmetrical antitoxin binding.
Nat Commun, 14, 2023
|
|
4WIV
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a novel inhibitor UMB32 (N-TERT-BUTYL-2-[4-(3,5-DIMETHYL-1,2-OXAZOL-4-YL) PHENYL]IMIDAZO[1,2-A]PYRAZIN-3-AMINE) | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, GLYCEROL, ... | | 著者 | Xu, X, Blacklow, S. | | 登録日 | 2014-09-26 | | 公開日 | 2014-10-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Biased multicomponent reactions to develop novel bromodomain inhibitors.
J.Med.Chem., 57, 2014
|
|
1TER
 
 | |
6WVT
 
 | | Structural basis of alphaE-catenin - F-actin catch bond behavior | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Xu, X.P, Pokutta, S, Torres, M, Swift, M.F, Hanein, D, Volkmann, N, Weis, W.I. | | 登録日 | 2020-05-06 | | 公開日 | 2020-10-07 | | 実験手法 | ELECTRON MICROSCOPY (3.56 Å) | | 主引用文献 | Structural basis of alpha E-catenin-F-actin catch bond behavior.
Elife, 9, 2020
|
|
6UWT
 
 | |
6UWR
 
 | |
4ZSO
 
 | | Crystal structure of a complex between B7-H6, a tumor cell ligand for natural cytotoxicity receptor NKp30, and an inhibitory antibody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | 著者 | Xu, X, Li, Y, Mariuzza, R.A. | | 登録日 | 2015-05-13 | | 公開日 | 2016-05-18 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of a complex between B7-H6, a tumor cell ligand for natural cytotoxicity receptor NKp30, and an inhibitory antibody
to be published
|
|
8AN4
 
 | | MenT1 toxin (rv0078a) from Mycobacterium tuberculosis H37Rv | | 分子名称: | Bacterial toxin | | 著者 | Xu, X, Usher, B, Gutierrez, C, Barriot, R, Arrowsmith, T.J, Han, X, Redder, P, Neyrolles, O, Blower, T.R, Genevaux, P. | | 登録日 | 2022-08-04 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | MenT nucleotidyltransferase toxins extend tRNA acceptor stems and can be inhibited by asymmetrical antitoxin binding.
Nat Commun, 14, 2023
|
|
4ZLP
 
 | |
8K9E
 
 | |
8K9F
 
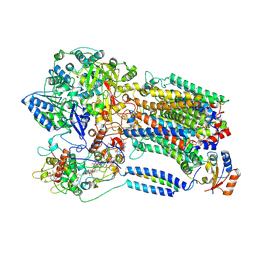 | |
7S0R
 
 | |
3VCB
 
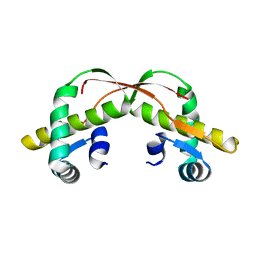 | | C425S mutant of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59 | | 分子名称: | RNA-directed RNA polymerase | | 著者 | Xu, X, Lou, Z, Ma, Y, Chen, X, Yang, Z, Tong, X, Zhao, Q, Xu, Y, Deng, H, Bartlam, M, Rao, Z. | | 登録日 | 2012-01-03 | | 公開日 | 2012-01-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59.
Plos One, 4, 2009
|
|
3VC8
 
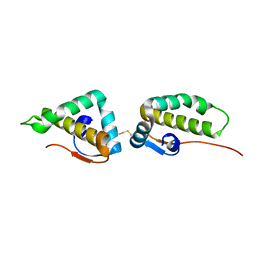 | | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59 | | 分子名称: | RNA-directed RNA polymerase | | 著者 | Xu, X, Lou, Z, Ma, Y, Chen, X, Yang, Z, Tong, X, Zhao, Q, Xu, Y, Deng, H, Bartlam, M, Rao, Z. | | 登録日 | 2012-01-03 | | 公開日 | 2012-01-11 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59.
Plos One, 4, 2009
|
|
7JTQ
 
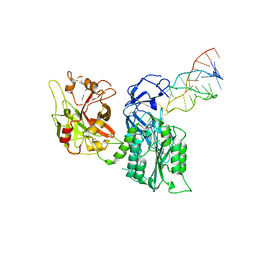 | |
