4AY8
 
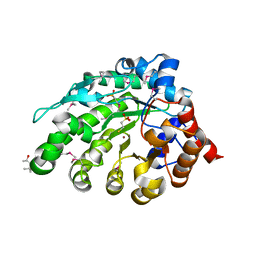 | | SeMet-derivative of a methyltransferase from M. mazei | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1-THIOETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Hoeppner, A, Thomas, F, Rueppel, A, Hensel, R, Blankenfeldt, W, Bayer, P, Faust, A. | | Deposit date: | 2012-06-18 | | Release date: | 2012-10-31 | | Last modified: | 2014-11-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Corrinoid:Coenzyme M Methyltransferase Mtaa from Methanosarcina Mazei
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4AY7
 
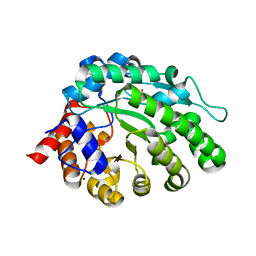 | | methyltransferase from Methanosarcina mazei | | Descriptor: | MAGNESIUM ION, METHYLCOBALAMIN: COENZYME M METHYLTRANSFERASE, ZINC ION | | Authors: | Hoeppner, A, Thomas, F, Rueppel, A, Hensel, R, Blankenfeld, W, Bayer, P, Faust, A. | | Deposit date: | 2012-06-18 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Corrinoid:Coenzyme M Methyltransferase Mtaa from Methanosarcina Mazei
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5N3U
 
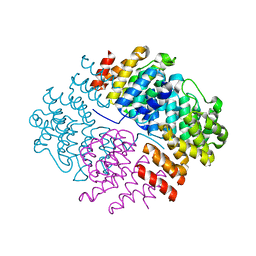 | | The structure of the complex of CpcE and CpcF of phycocyanin lyase from Nostoc sp. PCC7120 | | Descriptor: | Phycocyanobilin lyase subunit alpha, Phycocyanobilin lyase subunit beta | | Authors: | Hoeppner, A, Zhao, C, Xu, Q.-Z, Gaertner, W, Scheer, H, Zhao, K.-H. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-06 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structures and enzymatic mechanisms of phycobiliprotein lyases CpcE/F and PecE/F.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3JYP
 
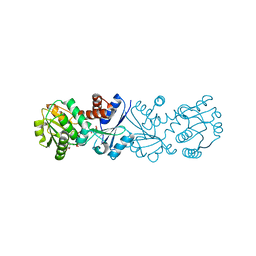 | | Quinate dehydrogenase from Corynebacterium glutamicum in complex with quinate and NADH | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Quinate/shikimate dehydrogenase | | Authors: | Hoeppner, A, Schomburg, D, Niefind, K. | | Deposit date: | 2009-09-22 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Enzyme-substrate complexes of the quinate/shikimate dehydrogenase from Corynebacterium glutamicum enable new insights in substrate and cofactor binding, specificity, and discrimination.
Biol.Chem., 394, 2013
|
|
3JYO
 
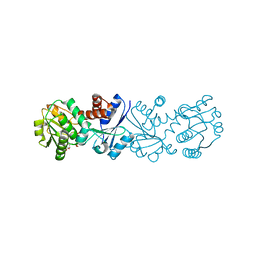 | | Quinate dehydrogenase from Corynebacterium glutamicum in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Quinate/shikimate dehydrogenase | | Authors: | Hoeppner, A, Niefind, K, Schomburg, D. | | Deposit date: | 2009-09-22 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Enzyme-substrate complexes of the quinate/shikimate dehydrogenase from Corynebacterium glutamicum enable new insights in substrate and cofactor binding, specificity, and discrimination.
Biol.Chem., 394, 2013
|
|
3JYQ
 
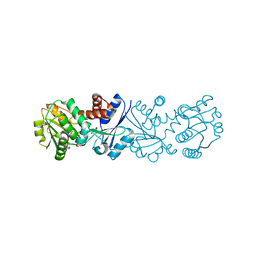 | | Quinate dehydrogenase from Corynebacterium glutamicum in complex with shikimate and NADH | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Quinate/shikimate dehydrogenase | | Authors: | Hoeppner, A, Schomburg, D, Niefind, K. | | Deposit date: | 2009-09-22 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Enzyme-substrate complexes of the quinate/shikimate dehydrogenase from Corynebacterium glutamicum enable new insights in substrate and cofactor binding, specificity, and discrimination.
Biol.Chem., 394, 2013
|
|
4Q5O
 
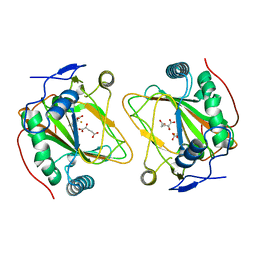 | | Crystal structure of EctD from S. alaskensis with 2-oxoglutarate and 5-hydroxyectoine | | Descriptor: | (4S,5S)-5-HYDROXY-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, 2-OXOGLUTARIC ACID, Ectoine hydroxylase, ... | | Authors: | Hoeppner, A, Widderich, N, Bremer, E, Smits, S.H. | | Deposit date: | 2014-04-17 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of the ectoine hydroxylase, a snapshot of the active site.
J.Biol.Chem., 289, 2014
|
|
4W1T
 
 | |
8BYK
 
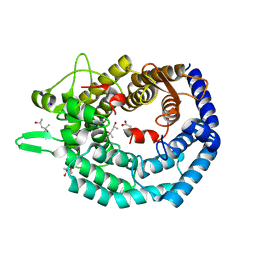 | | The structure of MadC from Clostridium maddingley reveals new insights into class I lanthipeptide cyclases | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Knospe, C.V, Kamel, M, Spitz, O, Hoeppner, A, Galle, S, Reiners, J, Kedrov, A, Smits, S.H, Schmitt, L. | | Deposit date: | 2022-12-13 | | Release date: | 2023-02-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of MadC from Clostridium maddingley reveals new insights into class I lanthipeptide cyclases.
Front Microbiol, 13, 2022
|
|
4UAH
 
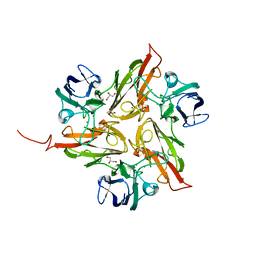 | |
4UAN
 
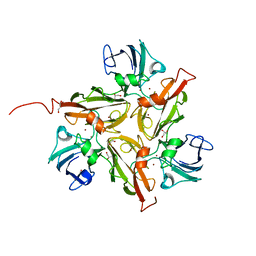 | |
4WTQ
 
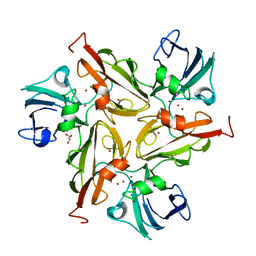 | |
4Y68
 
 | |
8B6E
 
 | | crystal structure of the DNA-binding short chromatophore-targeted protein sCTP-23166 from Paulinella chromatophora | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, sCTP-23166 | | Authors: | Macorano, L, Applegate, V, Hoeppner, A, Smits, S.H.J, Nowack, E.C.M. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | DNA-binding and protein structure of nuclear factors likely acting in genetic information processing in the Paulinella chromatophore.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6HRG
 
 | | Structure of Igni18, a novel metallo hydrolase from the hyperthermophilic archaeon Ignicoccus hospitalis KIN4/I | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, UPF0173 metal-dependent hydrolase Igni_1254, ... | | Authors: | Smits, S.H, Streit, W.R, Jaeger, K.E, Hoeppner, A. | | Deposit date: | 2018-09-26 | | Release date: | 2019-10-09 | | Last modified: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A promiscuous ancestral enzyme ́s structure unveils protein variable regions of the highly diverse metallo-beta-lactamase family.
Commun Biol, 4, 2021
|
|
8OR7
 
 | | Structure of a far-red induced allophycocyanin from Chroococcidiopsis thermalis sp. PCC 7203 | | Descriptor: | Allophycocyanin beta subunit apoprotein, PHYCOCYANOBILIN, POTASSIUM ION, ... | | Authors: | Zhou, L.J, Hoeppner, A, Wang, Y.Q, Zhao, K.H. | | Deposit date: | 2023-04-13 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic and biochemical analyses of a far-red allophycocyanin to address the mechanism of the super-red-shift.
Photosynth.Res., 2024
|
|
5M82
 
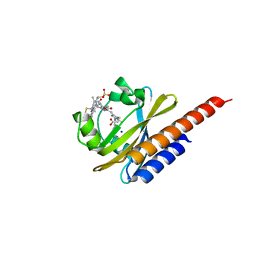 | | Three-dimensional structure of the photoproduct state of GAF3 from Slr1393 of Synechocystis sp. PCC6803 | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, PHYCOCYANOBILIN, SODIUM ION, ... | | Authors: | Xu, X.-L, Zhao, K.-H, Gaertner, W, Hoeppner, A. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural elements regulating the photochromicity in a cyanobacteriochrome
Proc.Natl.Acad.Sci.USA, 2020
|
|
5M85
 
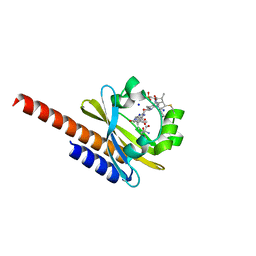 | | Three-dimensional structure of the intermediate state of GAF3 from Slr1393 of Synechocystis sp. PCC6803 | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHYCOCYANOBILIN, ... | | Authors: | Xu, X.-L, Zhao, K.-H, Gaertner, W, Hoeppner, A. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural elements regulating the photochromicity in a cyanobacteriochrome
Proc.Natl.Acad.Sci.USA, 2020
|
|
5DFY
 
 | |
5DCL
 
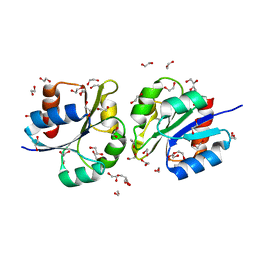 | | Structure of a lantibiotic response regulator: N terminal domain of the nisin resistance regulator NsrR | | Descriptor: | 1,2-ETHANEDIOL, PhoB family transcriptional regulator | | Authors: | Khosa, S, Kleinschrodt, D, Hoeppner, A, Smits, S.H. | | Deposit date: | 2015-08-24 | | Release date: | 2016-03-16 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structure of the Response Regulator NsrR from Streptococcus agalactiae, Which Is Involved in Lantibiotic Resistance.
Plos One, 11, 2016
|
|
5DCM
 
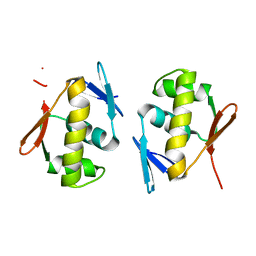 | | Structure of a lantibiotic response regulator: C-terminal domain of the nisin resistance regulator NsrR | | Descriptor: | PhoB family transcriptional regulator | | Authors: | Khosa, S, Kleinschrodt, D, Hoeppner, A, Smits, S.H.J. | | Deposit date: | 2015-08-24 | | Release date: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Response Regulator NsrR from Streptococcus agalactiae, Which Is Involved in Lantibiotic Resistance.
Plos One, 11, 2016
|
|
5DFX
 
 | |
5JVJ
 
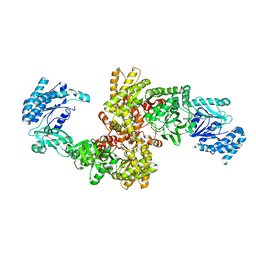 | | C4-type pyruvate phosphate dikinase: different conformational states of the nucleotide binding domain in the dimer | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE, Pyruvate, ... | | Authors: | Minges, A, Hoeppner, A, Groth, G. | | Deposit date: | 2016-05-11 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.898 Å) | | Cite: | Structural intermediates and directionality of the swiveling motion of Pyruvate Phosphate Dikinase.
Sci Rep, 7, 2017
|
|
5JVN
 
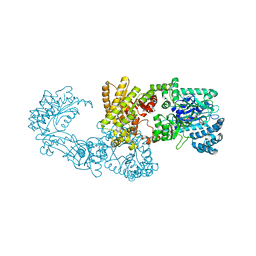 | | C3-type pyruvate phosphate dikinase: intermediate state of the central domain in the swiveling mechanism | | Descriptor: | 2'-Bromo-2'-deoxyadenosine 5'-[beta,gamma-imide]triphosphoric acid, MAGNESIUM ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Minges, A, Hoeppner, A, Groth, G. | | Deposit date: | 2016-05-11 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural intermediates and directionality of the swiveling motion of Pyruvate Phosphate Dikinase.
Sci Rep, 7, 2017
|
|
7PZE
 
 | | MademoiseLLE domain 2 of Rrm4 from Ustilago maydis | | Descriptor: | Chromosome 8, whole genome shotgun sequence | | Authors: | Devans, S, Schott-Verdugo, s, Muentjes, K, Olgeiser, L, Reiners, J, Schmitt, L, Hoeppner, A, Smits, S.H, Gohlke, H, Feldbruegge, M. | | Deposit date: | 2021-10-12 | | Release date: | 2022-06-15 | | Last modified: | 2022-12-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A MademoiseLLE domain binding platform links the key RNA transporter to endosomes.
Plos Genet., 18, 2022
|
|
