2JAE
 
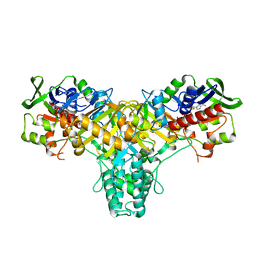 | | The structure of L-amino acid oxidase from Rhodococcus opacus in the unbound state | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID OXIDASE | | Authors: | Faust, A, Niefind, K, Hummel, W, Schomburg, D. | | Deposit date: | 2006-11-27 | | Release date: | 2007-01-30 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Structure of a Bacterial L-Amino Acid Oxidase from Rhodococcus Opacus Gives New Evidence for the Hydride Mechanism for Dehydrogenation.
J.Mol.Biol., 367, 2007
|
|
2JB3
 
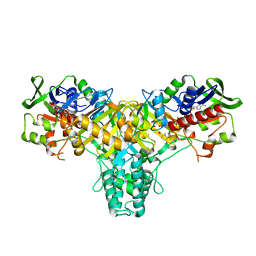 | | The structure of L-amino acid oxidase from Rhodococcus opacus in complex with o-aminobenzoate | | Descriptor: | 2-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID OXIDASE | | Authors: | Faust, A, Niefind, K, hummel, W, Schomburg, D. | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of a Bacterial L-Amino Acid Oxidase from Rhodococcus Opacus Gives New Evidence for the Hydride Mechanism for Dehydrogenation
J.Mol.Biol., 367, 2007
|
|
2JB2
 
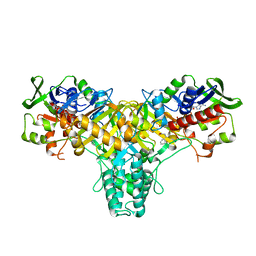 | | The structure of L-amino acid oxidase from Rhodococcus opacus in complex with L-phenylalanine. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID OXIDASE, PHENYLALANINE | | Authors: | Faust, A, Niefind, K, Hummel, W, Schomburg, D. | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Structure of a Bacterial L-Amino Acid Oxidase from Rhodococcus Opacus Gives New Evidence for the Hydride Mechanism for Dehydrogenation
J.Mol.Biol., 367, 2007
|
|
2JB1
 
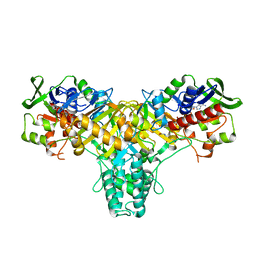 | | The L-amino acid oxidase from Rhodococcus opacus in complex with L- alanine | | Descriptor: | ALANINE, FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID OXIDASE | | Authors: | Faust, A, Niefind, K, Hummel, W, Schomburg, D. | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Structure of a Bacterial L-Amino Acid Oxidase from Rhodococcus Opacus Gives New Evidence for the Hydride Mechanism for Dehydrogenation
J.Mol.Biol., 367, 2007
|
|
3LQF
 
 | | Crystal structure of the short-chain dehydrogenase Galactitol-Dehydrogenase (GatDH) of Rhodobacter sphaeroides in complex with NAD and erythritol | | Descriptor: | Galactitol dehydrogenase, MAGNESIUM ION, MESO-ERYTHRITOL, ... | | Authors: | Carius, Y, Christian, H, Faust, A, Kornberger, P, Zander, U, Klink, B.U, Kohring, G.W, Giffhorn, F, Scheidig, A.J. | | Deposit date: | 2010-02-09 | | Release date: | 2010-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into substrate differentiation of the sugar-metabolizing enzyme galactitol dehydrogenase from Rhodobacter sphaeroides D.
J.Biol.Chem., 285, 2010
|
|
4AY8
 
 | | SeMet-derivative of a methyltransferase from M. mazei | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1-THIOETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Hoeppner, A, Thomas, F, Rueppel, A, Hensel, R, Blankenfeldt, W, Bayer, P, Faust, A. | | Deposit date: | 2012-06-18 | | Release date: | 2012-10-31 | | Last modified: | 2014-11-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Corrinoid:Coenzyme M Methyltransferase Mtaa from Methanosarcina Mazei
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4AY7
 
 | | methyltransferase from Methanosarcina mazei | | Descriptor: | MAGNESIUM ION, METHYLCOBALAMIN: COENZYME M METHYLTRANSFERASE, ZINC ION | | Authors: | Hoeppner, A, Thomas, F, Rueppel, A, Hensel, R, Blankenfeld, W, Bayer, P, Faust, A. | | Deposit date: | 2012-06-18 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Corrinoid:Coenzyme M Methyltransferase Mtaa from Methanosarcina Mazei
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2XTS
 
 | | Crystal Structure of the Sulfane Dehydrogenase SoxCD from Paracoccus pantotrophus | | Descriptor: | CALCIUM ION, COBALT (II) ION, CYTOCHROME, ... | | Authors: | Zander, U, Faust, A, Klink, B.U, de Sanctis, D, Panjikar, S, Quentmeier, A, Bardischewski, F, Friedrich, C.G, Scheidig, A.J. | | Deposit date: | 2010-10-12 | | Release date: | 2010-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural Basis for the Oxidation of Protein-Bound Sulfur by the Sulfur Cycle Molybdohemo-Enzyme Sulfane Dehydrogenase Soxcd.
J.Biol.Chem., 286, 2011
|
|
5NMW
 
 | | Crystal Structure of the pyrrolizidine alkaloid N-oxygenase from Zonocerus variegatus in complex with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase, MAGNESIUM ION | | Authors: | Scheidig, A, Kubitza, C, Faust, A, Ober, D. | | Deposit date: | 2017-04-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of pyrrolizidine alkaloid N-oxygenase from the grasshopper Zonocerus variegatus.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5NMX
 
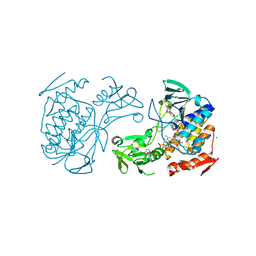 | | Crystal Structure of the pyrrolizidine alkaloid N-oxygenase from Zonocerus variegatus in complex with FAD and NADP+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase, MAGNESIUM ION, ... | | Authors: | Scheidig, A, Kubitza, C, Faust, A, Ober, D. | | Deposit date: | 2017-04-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of pyrrolizidine alkaloid N-oxygenase from the grasshopper Zonocerus variegatus.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4KOA
 
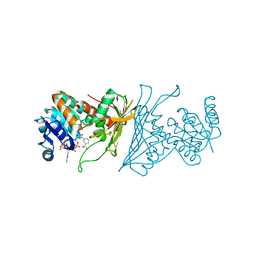 | | Crystal Structure Analysis of 1,5-anhydro-D-fructose reductase from Sinorhizobium meliloti | | Descriptor: | 1,5-anhydro-D-fructose reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Schu, M, Faust, A, Stosik, B, Kohring, G.-W, Giffhorn, F, Scheidig, A.J. | | Deposit date: | 2013-05-11 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The structure of substrate-free 1,5-anhydro-D-fructose reductase from Sinorhizobium meliloti 1021 reveals an open enzyme conformation.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
2WDZ
 
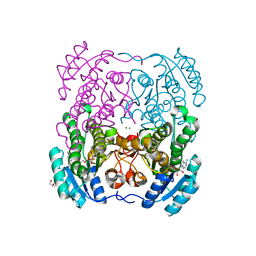 | | Crystal structure of the short chain dehydrogenase Galactitol- Dehydrogenase (GatDH) of Rhodobacter sphaeroides in complex with NAD+ and 1,2-Pentandiol | | Descriptor: | (2S)-pentane-1,2-diol, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Carius, Y, Christian, H, Faust, A, Kornberger, P, Kohring, G.W, Giffhorn, F, Scheidig, A.J. | | Deposit date: | 2009-03-27 | | Release date: | 2010-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insight Into Substrate Differentiation of the Sugar-Metabolizing Enzyme Galactitol Dehydrogenase from Rhodobacter Sphaeroides D.
J.Biol.Chem., 285, 2010
|
|
2WSB
 
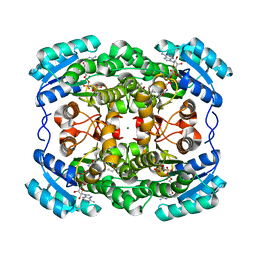 | | Crystal structure of the short-chain dehydrogenase Galactitol- Dehydrogenase (GatDH) of Rhodobacter sphaeroides in complex with NAD | | Descriptor: | GALACTITOL DEHYDROGENASE, MAGNESIUM ION, N-PROPANOL, ... | | Authors: | Carius, Y, Christian, H, Faust, A, Kornberger, P, Kohring, G.W, Giffhorn, F, Scheidig, A.J. | | Deposit date: | 2009-09-04 | | Release date: | 2010-04-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Insight Into Substrate Differentiation of the Sugar-Metabolizing Enzyme Galactitol Dehydrogenase from Rhodobacter Sphaeroides D.
J.Biol.Chem., 285, 2010
|
|
5OJG
 
 | | Crystal structure of the dehydrogenase/reductase SDR family member 4 (DHRS4) from Caenorhabditis elegans | | Descriptor: | Dehydrogenase/reductase SDR family member 4, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, butane-2,3-dione | | Authors: | Scheidig, A.J, Faust, A, Ebert, B, Maser, E, Kisiela, M. | | Deposit date: | 2017-07-21 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and catalytic characterization of the dehydrogenase/reductase SDR family member 4 (DHRS4) from Caenorhabditis elegans.
FEBS J., 285, 2018
|
|
5OJI
 
 | | Crystal structure of the dehydrogenase/reductase SDR family member 4 (DHRS4) from Caenorhabditis elegans | | Descriptor: | Dehydrogenase/reductase SDR family member 4, ISATIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Scheidig, A.J, Faust, A, Ebert, B, Maser, E, Kisiela, M. | | Deposit date: | 2017-07-21 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure and catalytic characterization of the dehydrogenase/reductase SDR family member 4 (DHRS4) from Caenorhabditis elegans.
FEBS J., 285, 2018
|
|
