6ZS5
 
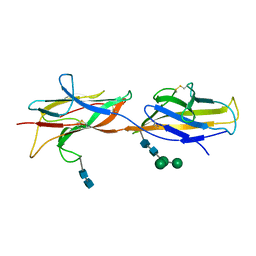 | | 3.5 A cryo-EM structure of human uromodulin filament core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stanisich, J.J, Zyla, D, Afanasyev, P, Xu, J, Pilhofer, M, Boeringer, D, Glockshuber, R. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism.
Elife, 9, 2020
|
|
6PWW
 
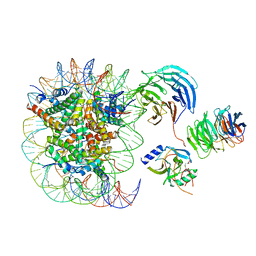 | | Cryo-EM structure of MLL1 in complex with RbBP5 and WDR5 bound to the nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
6ZYA
 
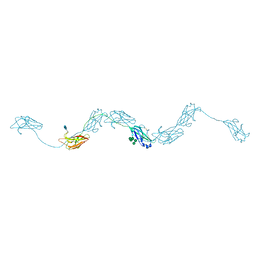 | | Extended human uromodulin filament core at 3.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stanisich, J.J, Zyla, D, Afanasyev, P, Xu, J, Pilhofer, M, Boeringer, D, Glockshuber, R. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism.
Elife, 9, 2020
|
|
6PWX
 
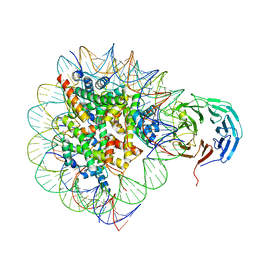 | | Cryo-EM structure of RbBP5 bound to the nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
5Y5E
 
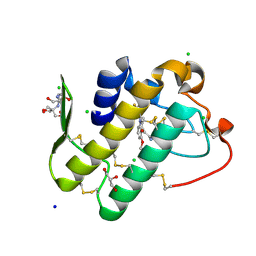 | | Crystal structure of phospholipase A2 with inhibitor | | Descriptor: | 2-[2-methyl-3-oxamoyl-1-[[2-(trifluoromethyl)phenyl]methyl]indol-4-yl]oxyethanoic acid, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-08-08 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2group IIE
Sci Rep, 7, 2017
|
|
5WZT
 
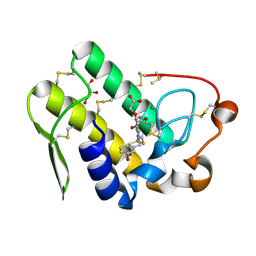 | | Crystal structure of human secreted phospholipase A2 group IIE with Compound 14 | | Descriptor: | 2-[1-[(3-bromophenyl)methyl]-2-methyl-3-oxamoyl-indol-4-yl]oxyethanoic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
5WZO
 
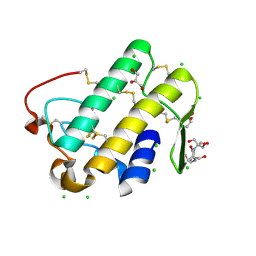 | | Crystal structure of human secreted phospholipase A2 group IIE, crystallized with calcium | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
5WZU
 
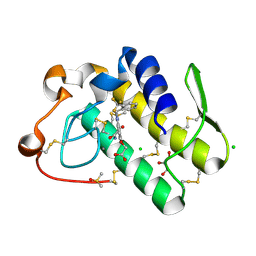 | | Crystal structure of human secreted phospholipase A2 group IIE with Compound 24 | | Descriptor: | 2-[2-methyl-3-oxamoyl-1-[[2-(trifluoromethyl)phenyl]methyl]indol-4-yl]oxyethanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
5WZW
 
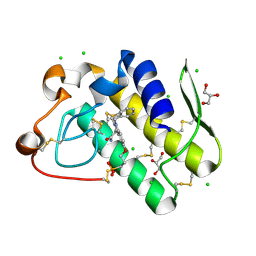 | | Crystal structure of human secreted phospholipase A2 group IIE with LY311727 | | Descriptor: | (3-{[3-(2-amino-2-oxoethyl)-1-benzyl-2-ethyl-1H-indol-5-yl]oxy}propyl)phosphonic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
5WZM
 
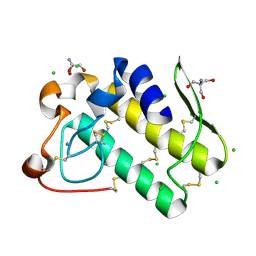 | | Crystal structure of human secreted phospholipase A2 group IIE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
5WZS
 
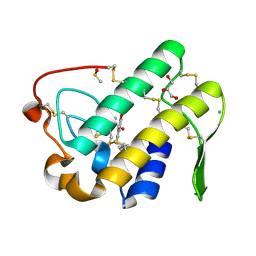 | | Crystal structure of human secreted phospholipase A2 group IIE with Compound 8 | | Descriptor: | 2-[2-methyl-1-(naphthalen-1-ylmethyl)-3-oxamoyl-indol-4-yl]oxyethanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
5WZV
 
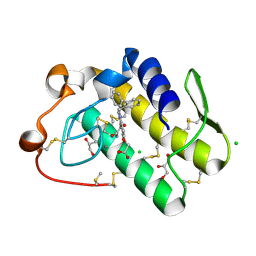 | | Crystal structure of human secreted phospholipase A2 group IIE with Me-indoxam | | Descriptor: | 2-[2-methyl-3-oxamoyl-1-[(2-phenylphenyl)methyl]indol-4-yl]oxyethanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
8SHJ
 
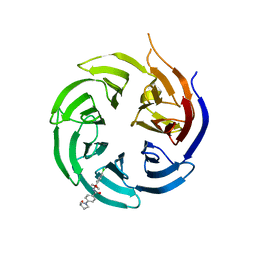 | | Crystal structure of the WD-repeat domain of human WDR91 in complex with MR45279 | | Descriptor: | N-[3-(4-chlorophenyl)oxetan-3-yl]-4-[(3S)-3-hydroxypyrrolidin-1-yl]benzamide, WD repeat-containing protein 91 | | Authors: | Ahmad, H, Zeng, H, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Xu, J, Feng, J.W, Brown, P.J, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-14 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery of a First-in-Class Small-Molecule Ligand for WDR91 Using DNA-Encoded Chemical Library Selection Followed by Machine Learning.
J.Med.Chem., 66, 2023
|
|
1QJB
 
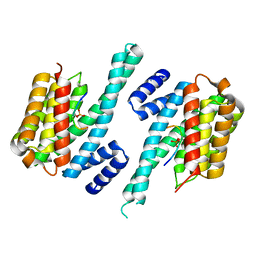 | | 14-3-3 ZETA/PHOSPHOPEPTIDE COMPLEX (MODE 1) | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, PHOSPHOPEPTIDE | | Authors: | Rittinger, K, Budman, J, Xu, J, Volinia, S, Cantley, L.C, Smerdon, S.J, Gamblin, S.J, Yaffe, M.B. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of 14-3-3 Phosphopeptide Complexes Identifies a Dual Role for the Nuclear Export Signal of 14-3-3 in Ligand Binding
Mol.Cell, 4, 1999
|
|
1QJA
 
 | | 14-3-3 ZETA/PHOSPHOPEPTIDE COMPLEX (MODE 2) | | Descriptor: | 14-3-3 PROTEIN ZETA, PHOSPHOPEPTIDE | | Authors: | Rittinger, K, Budman, J, Xu, J, Volinia, S, Cantley, L.C, Smerdon, S.J, Gamblin, S.J, Yaffe, M.B. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of 14-3-3 Phosphopeptide Complexes Identifies a Dual Role for the Nuclear Export Signal of 14-3-3 in Ligand Binding
Mol.Cell, 4, 1999
|
|
5XDZ
 
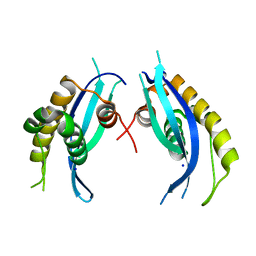 | | Crystal structure of zebrafish SNX25 PX domain | | Descriptor: | CHLORIDE ION, Cellular trafficking protein, SODIUM ION | | Authors: | Su, K, Zhang, Y, Xu, J, Liu, J. | | Deposit date: | 2017-03-30 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the PX domain of SNX25 reveals a novel phospholipid recognition model by dimerization in the PX domain
FEBS Lett., 591, 2017
|
|
2N1T
 
 | | Dynamic binding mode of a synaptotagmin-1-SNARE complex in solution | | Descriptor: | Synaptosomal-associated protein 25, Synaptotagmin-1, Syntaxin-1A, ... | | Authors: | Brewer, K, Bacaj, T, Cavalli, A, Camilloni, C, Swarbrick, J, Liu, J, Zhou, A, Zhou, P, Barlow, N, Xu, J, Seven, A, Prinslow, E, Voleti, R, Haussinger, D, Bonvin, A, Tomchick, D, Vendruscolo, M, Graham, B, Sudhof, T, Rizo, J. | | Deposit date: | 2015-04-21 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dynamic binding mode of a Synaptotagmin-1-SNARE complex in solution.
Nat.Struct.Mol.Biol., 22, 2015
|
|
200L
 
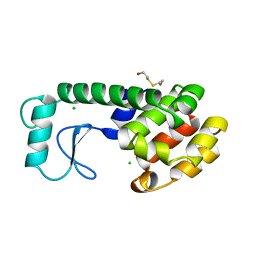 | | THERMODYNAMIC AND STRUCTURAL COMPENSATION IN "SIZE-SWITCH" CORE-REPACKING VARIANTS OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Baldwin, E, Xu, J, Hajiseyedjavadi, O, Matthews, B.W. | | Deposit date: | 1995-11-06 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Thermodynamic and structural compensation in "size-switch" core repacking variants of bacteriophage T4 lysozyme.
J.Mol.Biol., 259, 1996
|
|
4NPK
 
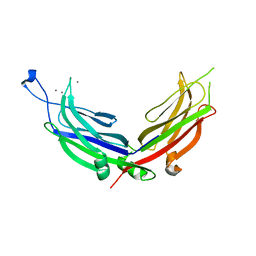 | | Extended-Synaptotagmin 2, C2A- and C2B-domains, calcium bound | | Descriptor: | CALCIUM ION, Extended synaptotagmin-2 | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2013-11-21 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Structure and ca(2+)-binding properties of the tandem c2 domains of e-syt2.
Structure, 22, 2014
|
|
4NPJ
 
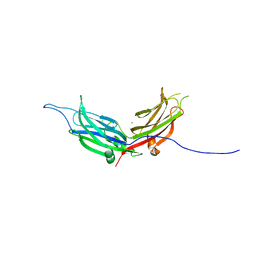 | | Extended-Synaptotagmin 2, C2A- and C2B-domains | | Descriptor: | ACETATE ION, CHLORIDE ION, Extended synaptotagmin-2, ... | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2013-11-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structure and ca(2+)-binding properties of the tandem c2 domains of e-syt2.
Structure, 22, 2014
|
|
4DGP
 
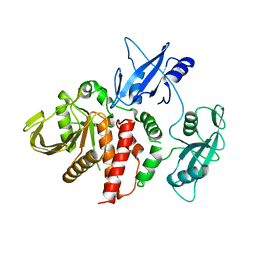 | | The wild-type Src homology 2 (SH2)-domain containing protein tyrosine phosphatase-2 (SHP2) | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Yu, Z.H, Xu, J, Walls, C.D, Chen, L, Zhang, S, Wu, L, Wang, L.N, Liu, S.J, Zhang, Z.Y. | | Deposit date: | 2012-01-26 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Insights into LEOPARD Syndrome-Associated SHP2 Mutations.
J.Biol.Chem., 288, 2013
|
|
7KH0
 
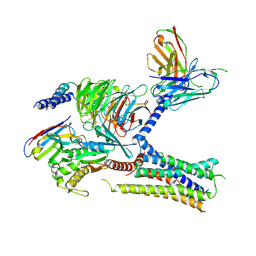 | | Cryo-EM structure of the human arginine vasopressin AVP-vasopressin receptor V2R-Gs signaling complex | | Descriptor: | Arg-vasopressin, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, L, Xu, J, Gao, S, Sun, D, Liu, H, Liu, Z, Du, Y, Zhang, C. | | Deposit date: | 2020-10-19 | | Release date: | 2021-05-26 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of the AVP-vasopressin receptor 2-G s signaling complex.
Cell Res., 31, 2021
|
|
4DGX
 
 | | LEOPARD Syndrome-Associated SHP2/Y279C mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Yu, Z.H, Xu, J, Walls, C.D, Chen, L, Zhang, S, Wu, L, Wang, L.N, Liu, S.J, Zhang, Z.Y. | | Deposit date: | 2012-01-27 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Insights into LEOPARD Syndrome-Associated SHP2 Mutations.
J.Biol.Chem., 288, 2013
|
|
3QI4
 
 | | Crystal structure of PDE9A(Q453E) in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Hou, J, Xu, J, Liu, M, Zhao, R, Lou, H, Ke, H. | | Deposit date: | 2011-01-26 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural asymmetry of phosphodiesterase-9, potential protonation of a glutamic Acid, and role of the invariant glutamine.
Plos One, 6, 2011
|
|
3N3Z
 
 | | Crystal structure of PDE9A (E406A) mutant in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, CHLORIDE ION, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, ... | | Authors: | Hou, J, Luo, H.-B, Chen, Y, Xu, J, Zhao, R, Zou, L. | | Deposit date: | 2010-05-21 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of PDE9A (E406A) mutation in complex with IBMX
To be Published
|
|
