4JN4
 
 | | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaK, GLYCEROL, ... | | Authors: | Qi, R, Sarbeng, E.B, Liu, Q, Le, K.Q, Xu, X, Xu, H, Yang, J, Wong, J.L, Vorvis, C, Hendrickson, W.A, Zhou, L, Liu, Q. | | Deposit date: | 2013-03-14 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6YMB
 
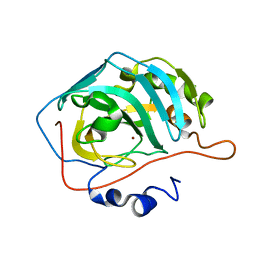 | | MicroED structure of human carbonic anhydrase II | | Descriptor: | ZINC ION, carbonic anhydrase 2 | | Authors: | Clabbers, M.T.B, Fisher, S.Z, Coincon, M, Zou, X, Xu, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Visualizing drug binding interactions using microcrystal electron diffraction.
Commun Biol, 3, 2020
|
|
6YMA
 
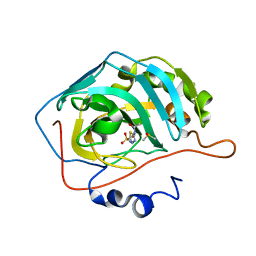 | | MicroED structure of acetazolamide-bound human carbonic anhydrase II | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, DIMETHYL SULFOXIDE, ZINC ION, ... | | Authors: | Clabbers, M.T.B, Fisher, S.Z, Coincon, M, Zou, X, Xu, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Visualizing drug binding interactions using microcrystal electron diffraction.
Commun Biol, 3, 2020
|
|
4JNE
 
 | | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Hsp70 CHAPERONE DnaK, ... | | Authors: | Qi, R, Sarbeng, E.B, Liu, Q, Le, K.Q, Xu, X, Xu, H, Yang, J, Wong, J.L, Vorvis, C, Hendrickson, W.A, Zhou, L, Liu, Q. | | Deposit date: | 2013-03-15 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP.
Nat.Struct.Mol.Biol., 20, 2013
|
|
8X7A
 
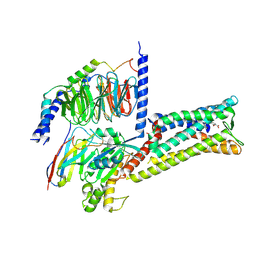 | | Treprostinil bound Prostacyclin Receptor G protein complex | | Descriptor: | 2-[[(1~{R},2~{R},3~{a}~{S},9~{a}~{S})-2-oxidanyl-1-[(3~{S})-3-oxidanyloctyl]-2,3,3~{a},4,9,9~{a}-hexahydro-1~{H}-cyclopenta[g]naphthalen-5-yl]oxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.J, Jin, S, Zhang, H, Xu, Y, Hu, W, Jiang, Y, Chen, C, Wang, D.W, Xu, H.E, Wu, C. | | Deposit date: | 2023-11-23 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Molecular recognition and activation of the prostacyclin receptor by anti-pulmonary arterial hypertension drugs.
Sci Adv, 10, 2024
|
|
3NMH
 
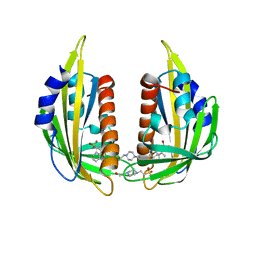 | | Crystal structure of the abscisic receptor PYL2 in complex with pyrabactin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | Deposit date: | 2010-06-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
8X17
 
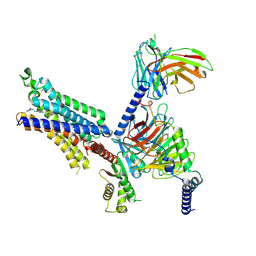 | | Cryo-EM structure of adenosine receptor A3AR bound to CF102 | | Descriptor: | Adenosine receptor A3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cai, H, Xu, Y, Xu, H.E. | | Deposit date: | 2023-11-06 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Cryo-EM structures of adenosine receptor A 3 AR bound to selective agonists.
Nat Commun, 15, 2024
|
|
8X16
 
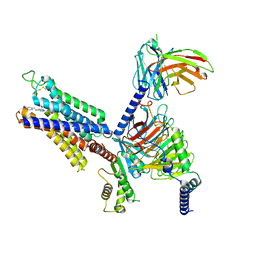 | | Cryo-EM structure of adenosine receptor A3AR bound to CF101 | | Descriptor: | Adenosine receptor A3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cai, H, Xu, Y, Xu, H.E. | | Deposit date: | 2023-11-06 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structures of adenosine receptor A 3 AR bound to selective agonists.
Nat Commun, 15, 2024
|
|
3NMV
 
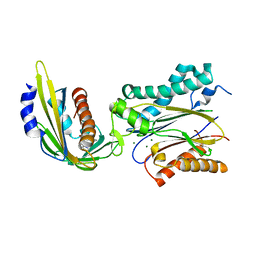 | | Crystal structure of pyrabactin-bound abscisic acid receptor PYL2 mutant A93F in complex with type 2C protein phosphatase ABI2 | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | Deposit date: | 2010-06-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NMT
 
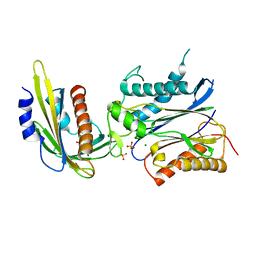 | | Crystal structure of pyrabactin bound abscisic acid receptor PYL2 mutant A93F in complex with type 2C protein phosphatase HAB1 | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | Deposit date: | 2010-06-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4LGB
 
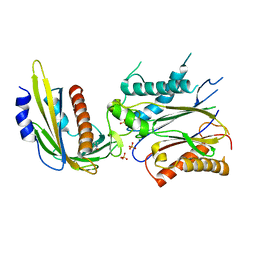 | | ABA-mimicking ligand N-(1-METHYL-2-OXO-1,2,3,4-TETRAHYDROQUINOLIN-6-YL)-1-(4-METHYLPHENYL)METHANESULFONAMIDE in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | Abscisic acid receptor PYL2, MAGNESIUM ION, N-(1-methyl-2-oxo-1,2,3,4-tetrahydroquinolin-6-yl)-1-(4-methylphenyl)methanesulfonamide, ... | | Authors: | Zhou, X.E, Gao, M, Liu, X, Zhang, Y, Xue, X, Melcher, K, Gao, P, Wang, F, Zeng, L, Zhao, Y, Zhao, Y, Deng, P, Zhong, D, Zhu, J.-K, Xu, Y, Xu, H.E. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants.
Cell Res., 23, 2013
|
|
4LG5
 
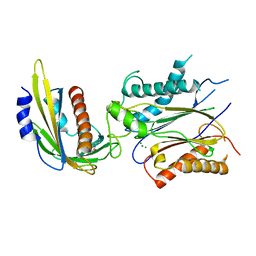 | | ABA-mimicking ligand QUINABACTIN in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | Abscisic acid receptor PYL2, MAGNESIUM ION, Protein phosphatase 2C 16, ... | | Authors: | Zhou, X.E, Gao, M, Liu, X, Zhang, Y, Xue, X, Melcher, K, Gao, P, Wang, F, Zeng, L, Zhao, Y, Zhao, Y, Deng, P, Zhong, D, Zhu, J.-K, Xu, Y, Xu, H.E. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants.
Cell Res., 23, 2013
|
|
4LGA
 
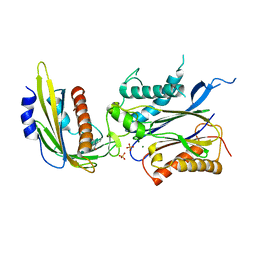 | | ABA-mimicking ligand N-(2-OXO-1-PROPYL-1,2,3,4-TETRAHYDROQUINOLIN-6-YL)-1-PHENYLMETHANESULFONAMIDE in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | Abscisic acid receptor PYL2, MAGNESIUM ION, N-(2-oxo-1-propyl-1,2,3,4-tetrahydroquinolin-6-yl)-1-phenylmethanesulfonamide, ... | | Authors: | Zhou, X.E, Gao, M, Liu, X, Zhang, Y, Xue, X, Melcher, K, Gao, P, Wang, F, Zeng, L, Zhao, Y, Zhao, Y, Deng, P, Zhong, D, Zhu, J.-K, Xu, Y, Xu, H.E. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants.
Cell Res., 23, 2013
|
|
3NMN
 
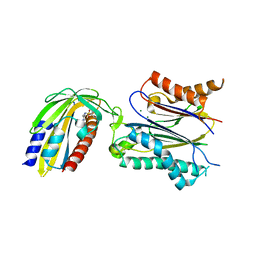 | | Crystal structure of pyrabactin-bound abscisic acid receptor PYL1 in complex with type 2C protein phosphatase ABI1 | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL1, MAGNESIUM ION, ... | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | Deposit date: | 2010-06-22 | | Release date: | 2010-08-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NMP
 
 | | Crystal structure of the abscisic receptor PYL2 mutant A93F in complex with pyrabactin | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | Deposit date: | 2010-06-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1GCB
 
 | | GAL6, YEAST BLEOMYCIN HYDROLASE DNA-BINDING PROTEASE (THIOL) | | Descriptor: | GAL6 HG (EMTS) DERIVATIVE, GLYCEROL, MERCURY (II) ION, ... | | Authors: | Joshua-Tor, L, Xu, H.E, Johnston, S.A, Rees, D.C. | | Deposit date: | 1995-07-18 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a conserved protease that binds DNA: the bleomycin hydrolase, Gal6.
Science, 269, 1995
|
|
2P54
 
 | | a crystal structure of PPAR alpha bound with SRC1 peptide and GW735 | | Descriptor: | 2-METHYL-2-(4-{[({4-METHYL-2-[4-(TRIFLUOROMETHYL)PHENYL]-1,3-THIAZOL-5-YL}CARBONYL)AMINO]METHYL}PHENOXY)PROPANOIC ACID, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor alpha | | Authors: | Xu, R.X, Xu, H.E, Sierra, M.L, Montana, V.G, Lambert, M.H, Pianetti, P.M. | | Deposit date: | 2007-03-14 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Substituted 2-[(4-Aminomethyl)phenoxy]-2-methylpropionic Acid PPAR Agonists. 1.Discovery of a Novel Series of Potent HDLc Raising Agents.
J.Med.Chem., 50, 2007
|
|
5TO5
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-10-16 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
5CL1
 
 | | Complex structure of Norrin with human Frizzled 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Frizzled-4, Maltose-binding periplasmic protein,Norrin | | Authors: | Wang, Z, Ke, J, Shen, G, Cheng, Z, Xu, H.E, Xu, W. | | Deposit date: | 2015-07-16 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of the Norrin-Frizzled 4 interaction.
Cell Res., 25, 2015
|
|
4RED
 
 | | Crystal structure of human AMPK alpha1 KD-AID with K43A mutation | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1 | | Authors: | Zhou, X.E, Ke, J, Li, X, Wang, L, Gu, X, de Waal, P.W, Tan, M.H.E, Wang, D, Wu, D, Xu, H.E, Melcher, K. | | Deposit date: | 2014-09-22 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of AMPK regulation by adenine nucleotides and glycogen.
Cell Res., 25, 2015
|
|
4RER
 
 | | Crystal structure of the phosphorylated human alpha1 beta2 gamma1 holo-AMPK complex bound to AMP and cyclodextrin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, ... | | Authors: | Zhou, X.E, Ke, J, Li, X, Wang, L, Gu, X, de Waal, P.W, Tan, M.H.E, Wang, D, Wu, D, Xu, H.E, Melcher, K. | | Deposit date: | 2014-09-23 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.047 Å) | | Cite: | Structural basis of AMPK regulation by adenine nucleotides and glycogen.
Cell Res., 25, 2015
|
|
5GW8
 
 | | Crystal structure of a putative DAG-like lipase (MgMDL2) from Malassezia globosa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Xu, J, Xu, H, Liu, J. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Malassezia globosa MgMDL2 lipase: Crystal structure and rational modification of substrate specificity.
Biochem. Biophys. Res. Commun., 488, 2017
|
|
5TVB
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-11-08 | | Release date: | 2017-09-20 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
5DGY
 
 | | Crystal structure of rhodopsin bound to visual arrestin | | Descriptor: | Endolysin,Rhodopsin,S-arrestin | | Authors: | Zhou, X.E, Gao, X, Kang, Y, He, Y, de Waal, P.W, Suino-Powell, K.M, Wang, M, Melcher, K, Xu, H.E. | | Deposit date: | 2015-08-28 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (7.7 Å) | | Cite: | X-ray laser diffraction for structure determination of the rhodopsin-arrestin complex.
Sci Data, 3, 2016
|
|
8XIQ
 
 | | Structure of L796778-SSTR3 G protein complex | | Descriptor: | G-alpha i, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Xu, Y, Xu, H.E, Zhuang, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Selective ligand recognition and activation of somatostatin receptors SSTR1 and SSTR3
To Be Published
|
|
