7XW1
 
 | |
1KMF
 
 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT ILE-A2-ALLO-ILE, HIS-B10-ASP, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | 分子名称: | Insulin | | 著者 | Xu, B, Hua, Q.X, Nakagawa, S.H, Jia, W, Chu, Y.C, Katsoyannis, P.G, Weiss, M.A. | | 登録日 | 2001-12-14 | | 公開日 | 2002-01-09 | | 最終更新日 | 2021-10-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Chiral mutagenesis of insulin's hidden receptor-binding surface: structure of an allo-isoleucine(A2) analogue.
J.Mol.Biol., 316, 2002
|
|
1K3M
 
 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT ILE-A2-ALA, HIS-B10-ASP, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | 分子名称: | INSULIN | | 著者 | Xu, B, Hua, Q.-X, Nakagawa, S.H, Jia, W, Chu, Y.-C, Katsoyannis, P.G, Weiss, M.A. | | 登録日 | 2001-10-03 | | 公開日 | 2001-10-17 | | 最終更新日 | 2024-10-30 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A cavity-forming mutation in insulin induces segmental unfolding of a surrounding alpha-helix.
Protein Sci., 11, 2002
|
|
7YR5
 
 | |
4QSG
 
 | | Crystal structure of gas vesicle protein GvpF from Microcystis aeruginosa | | 分子名称: | Gas vesicle protein | | 著者 | Xu, B.Y, Dai, Y.N, Zhou, K, Ren, Y.M, Liu, Y.T, Chen, Y, Zhou, C.Z. | | 登録日 | 2014-07-03 | | 公開日 | 2014-11-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of the gas vesicle protein GvpF from the cyanobacterium Microcystis aeruginosa.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3OMZ
 
 | |
7FVS
 
 | | Crystal Structure of S. aureus gyrase in complex with 4-[[1-[(1-chloro-6,7-dihydro-5H-cyclopenta[c]pyridin-6-yl)methyl]azetidin-3-yl]methylamino]-6-fluorochromen-2-one | | 分子名称: | 4-{[(1-{[(6R)-1-chloro-6,7-dihydro-5H-cyclopenta[c]pyridin-6-yl]methyl}azetidin-3-yl)methyl]amino}-6-fluoro-2H-1-benzopyran-2-one, ACETATE ION, CHLORIDE ION, ... | | 著者 | Xu, B, Benz, J, Cumming, J.G, Kreis, L, Rudolph, M.G. | | 登録日 | 2023-04-18 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Discovery of a Series of Indane-Containing NBTIs with Activity against Multidrug-Resistant Gram-Negative Pathogens.
Acs Med.Chem.Lett., 14, 2023
|
|
7FVT
 
 | | Crystal Structure of S. aureus gyrase in complex with 6-[5-[2-[(4-chloro-2,3-dihydro-1H-inden-2-yl)methylamino]ethyl]-2-oxo-1,3-oxazolidin-3-yl]-4H-pyrido[3,2-b][1,4]oxazin-3-one | | 分子名称: | 6-{(5R)-5-[2-({[(2R)-4-chloro-2,3-dihydro-1H-inden-2-yl]methyl}amino)ethyl]-2-oxo-1,3-oxazolidin-3-yl}-2H-pyrido[3,2-b][1,4]oxazin-3(4H)-one, CHLORIDE ION, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | 著者 | Xu, B, Benz, J, Cumming, J.G, Rudolph, M.G. | | 登録日 | 2023-04-18 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.081 Å) | | 主引用文献 | Discovery of a Series of Indane-Containing NBTIs with Activity against Multidrug-Resistant Gram-Negative Pathogens.
Acs Med.Chem.Lett., 14, 2023
|
|
3JSD
 
 | | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus | | 分子名称: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | 著者 | Weiss, M.A, Wan, Z.L, Dodson, E.J, Liu, M, Xu, B, Hua, Q.X, Turkenburg, M, Whittingham, J, Nakagawa, S.H, Huang, K, Hu, S.Q, Jia, W.H, Wang, S.H, Brange, J, Whittaker, J, Arvan, P, Katsoyannis, P.G, Dodson, G.G. | | 登録日 | 2009-09-10 | | 公開日 | 2010-09-15 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus
To be Published
|
|
5WRQ
 
 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | 分子名称: | 5-[[2,4-bis(oxidanylidene)quinazolin-1-yl]methyl]-2-fluoranyl-N-[(3R)-1-(3-methylbutyl)pyrrolidin-3-yl]benzamide, Poly [ADP-ribose] polymerase 1 | | 著者 | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | 登録日 | 2016-12-03 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
5WS0
 
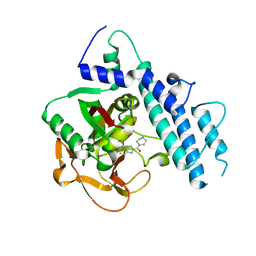 | | Structure of human PARP1 catalytic domain bound to a benzoimidazole inhibitor | | 分子名称: | 2-piperazin-1-ylcarbonyl-1H-benzimidazole-4-carboxamide, Poly [ADP-ribose] polymerase 1 | | 著者 | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | 登録日 | 2016-12-04 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of human PARP1 catalytic domain bound to a benzoimidazole inhibitor
To Be Published
|
|
5WS1
 
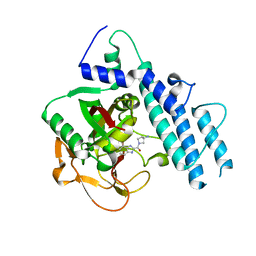 | | Structure of human PARP1 catalytic domain bound to a benzoimidazole inhibitor | | 分子名称: | 2-[(3R)-3-azanylpyrrolidin-1-yl]carbonyl-1H-benzimidazole-4-carboxamide, Poly [ADP-ribose] polymerase 1 | | 著者 | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | 登録日 | 2016-12-04 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of human PARP1 catalytic domain bound to a benzoimidazole inhibitor
To Be Published
|
|
5WTC
 
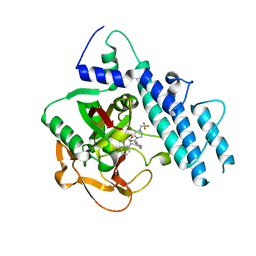 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | 分子名称: | 1-[[4-fluoranyl-3-[4-[2,2,2-tris(fluoranyl)ethyl]piperazin-1-yl]carbonyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1 | | 著者 | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | 登録日 | 2016-12-11 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
9BJN
 
 | | Cryo-EM of Azo-ffspy fiber | | 分子名称: | D-peptide ffspy | | 著者 | Zia, A, Guo, J, Xu, B, Wang, F. | | 登録日 | 2024-04-25 | | 公開日 | 2024-09-18 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Cell-Free Nonequilibrium Assembly for Hierarchical Protein/Peptide Nanopillars.
J.Am.Chem.Soc., 146, 2024
|
|
9BJO
 
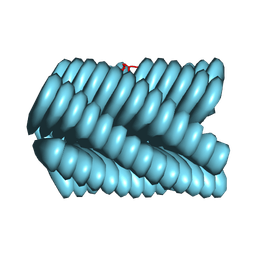 | | Cryo-EM of Azo-ffsy fiber | | 分子名称: | D-peptide ffsy | | 著者 | Zia, A, Guo, J, Xu, B, Wang, F. | | 登録日 | 2024-04-25 | | 公開日 | 2024-09-18 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Cell-Free Nonequilibrium Assembly for Hierarchical Protein/Peptide Nanopillars.
J.Am.Chem.Soc., 146, 2024
|
|
6LW2
 
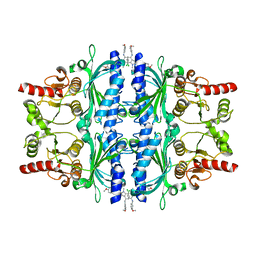 | | The N-arylsulfonyl-indole-2-carboxamide-based inhibitors against fructose-1,6-bisphosphatase | | 分子名称: | 7-chloranyl-4-[(3-methoxyphenyl)amino]-N-(4-methoxyphenyl)sulfonyl-1-methyl-indole-2-carboxamide, Fructose-1,6-bisphosphatase 1 | | 著者 | Wang, X.Y, Zhou, J, Xu, B.L. | | 登録日 | 2020-02-07 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Discovery of N -Arylsulfonyl-Indole-2-Carboxamide Derivatives as Potent, Selective, and Orally Bioavailable Fructose-1,6-Bisphosphatase Inhibitors-Design, Synthesis, In Vivo Glucose Lowering Effects, and X-ray Crystal Complex Analysis.
J.Med.Chem., 63, 2020
|
|
8FOF
 
 | |
6RPG
 
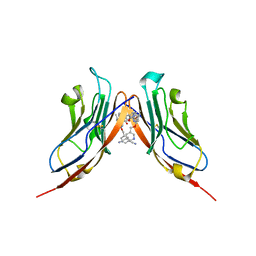 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with inhibitor | | 分子名称: | Programmed cell death 1 ligand 1, ~{N}-[2-[[4-[[3-[3-[[4-[(2-acetamidoethylamino)methyl]-5-[(5-cyanopyridin-3-yl)methoxy]-2-methyl-phenoxy]methyl]-2-methyl-phenyl]-2-methyl-phenyl]methoxy]-2-[(5-cyanopyridin-3-yl)methoxy]-5-methyl-phenyl]methylamino]ethyl]ethanamide | | 著者 | Magiera-Mularz, K, Basu, S, Yang, J, Xu, B, Skalniak, L, Musielak, B, Kholodovych, V, Holak, T.A, Hu, L. | | 登録日 | 2019-05-14 | | 公開日 | 2019-07-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Design, Synthesis, Evaluation, and Structural Studies ofC2-Symmetric Small Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
6X5I
 
 | |
8DST
 
 | |
9IUZ
 
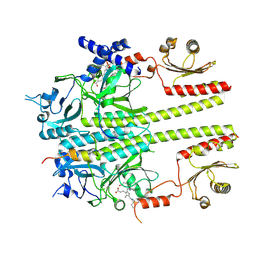 | | Constitutively active mutant(Y276H) of Arabidopsis phytochrome B(phyB) in complex with phytochrome-interacting factor 6(PIF6) | | 分子名称: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome B, Phytochrome-interacting factor 6 | | 著者 | Wang, Z, Wang, W, Zhao, D, Song, Y, Xu, B, Zhao, J, Wang, J. | | 登録日 | 2024-07-22 | | 公開日 | 2024-10-02 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.19 Å) | | 主引用文献 | Light-induced remodeling of phytochrome B enables signal transduction by phytochrome-interacting factor.
Cell, 2024
|
|
1LEW
 
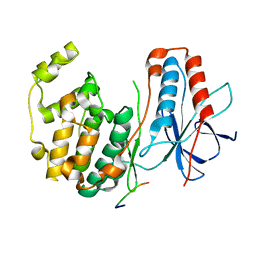 | | CRYSTAL STRUCTURE OF MAP KINASE P38 COMPLEXED TO THE DOCKING SITE ON ITS NUCLEAR SUBSTRATE MEF2A | | 分子名称: | Mitogen-activated protein kinase 14, Myocyte-specific enhancer factor 2A | | 著者 | Chang, C.-I, Xu, B.-E, Akella, R, Cobb, M.H, Goldsmith, E.J. | | 登録日 | 2002-04-10 | | 公開日 | 2002-07-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b.
Mol.Cell, 9, 2002
|
|
1LEZ
 
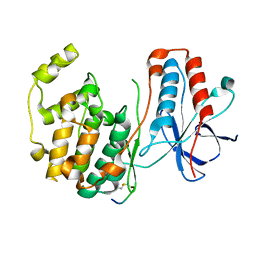 | | CRYSTAL STRUCTURE OF MAP KINASE P38 COMPLEXED TO THE DOCKING SITE ON ITS ACTIVATOR MKK3B | | 分子名称: | MAP kinase kinase 3b, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | 著者 | Chang, C.-I, Xu, B.-E, Akella, R, Cobb, M.H, Goldsmith, E.J. | | 登録日 | 2002-04-10 | | 公開日 | 2002-07-10 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b.
Mol.Cell, 9, 2002
|
|
8YB4
 
 | | Pfr conformer of Arabidopsis thaliana phytochrome B in complex with phytochrome-interacting factor 6 | | 分子名称: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, phytochrome B, phytochrome-interacting factor 6 | | 著者 | Wang, Z, Wang, W, Zhao, D, Song, Y, Xu, B, Zhao, J, Wang, J. | | 登録日 | 2024-02-11 | | 公開日 | 2024-10-02 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Light-induced remodeling of phytochrome B enables signal transduction by phytochrome-interacting factor.
Cell, 2024
|
|
2YOP
 
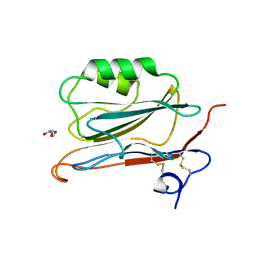 | | Long wavelength S-SAD structure of FAM3B PANDER | | 分子名称: | GLYCEROL, PROTEIN FAM3B | | 著者 | Johansson, P, Bernstrom, J, Gorman, T, Oster, L, Backstrom, S, Schweikart, F, Xu, B, Xue, Y, Holmberg Schiavone, L. | | 登録日 | 2012-10-26 | | 公開日 | 2013-01-30 | | 最終更新日 | 2013-02-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Fam3B Pander and Fam3C Ilei Represent a Distinct Class of Signaling Molecules with a Non-Cytokine-Like Fold.
Structure, 21, 2013
|
|
